flowchart TB ML(Machine learning) --> UL(Unsupervised learning) ML --> SL(Supervised learning) ML --> RL(Reinforcement learning) ML --> dots1(...) SL --> BC(binary classification) SL --> MCC(multi-class classification) SL --> MLC(multi-label classification) SL --> TTE(time-to-event analysis) SL --> REG(regression) SL --> dots2(...)
Model and Algorithm Evaluation in Supervised Machine Learning
Short Course at CEN 2023 Conference
Fraunhofer Institute for Digital Medicine MEVIS
2023-09-03
Introduction
Scope
- Important topics (50-80%):
- Core concepts
- Performance metrics
- Data splitting
- Statistical analysis
- Further considerations (20-50%)
- Practical aspects
- topics not covered in this course…
- Learning goal:
- be able to identify common pitfalls for your ML problem…
- … and find suitable (evaluation) solutions.
Housekeeping
- Slides & reproducible code
- Interactive summaries
- at the end of each section
- Questions
- at the end of each section
- online participants: may use the chat
Agenda
- Introduction \(\longrightarrow\)
- Core Concepts \(\longrightarrow\) \(\longrightarrow\)
- Performance metrics \(\longrightarrow\) \(\longrightarrow\)
- Break
- Data splitting \(\longrightarrow\) \(\longrightarrow\)
- Statistical analysis \(\longrightarrow\) \(\longrightarrow\)
- Practical aspects \(\longrightarrow\) \(\longrightarrow\)
- Wrap-up
Core Concepts
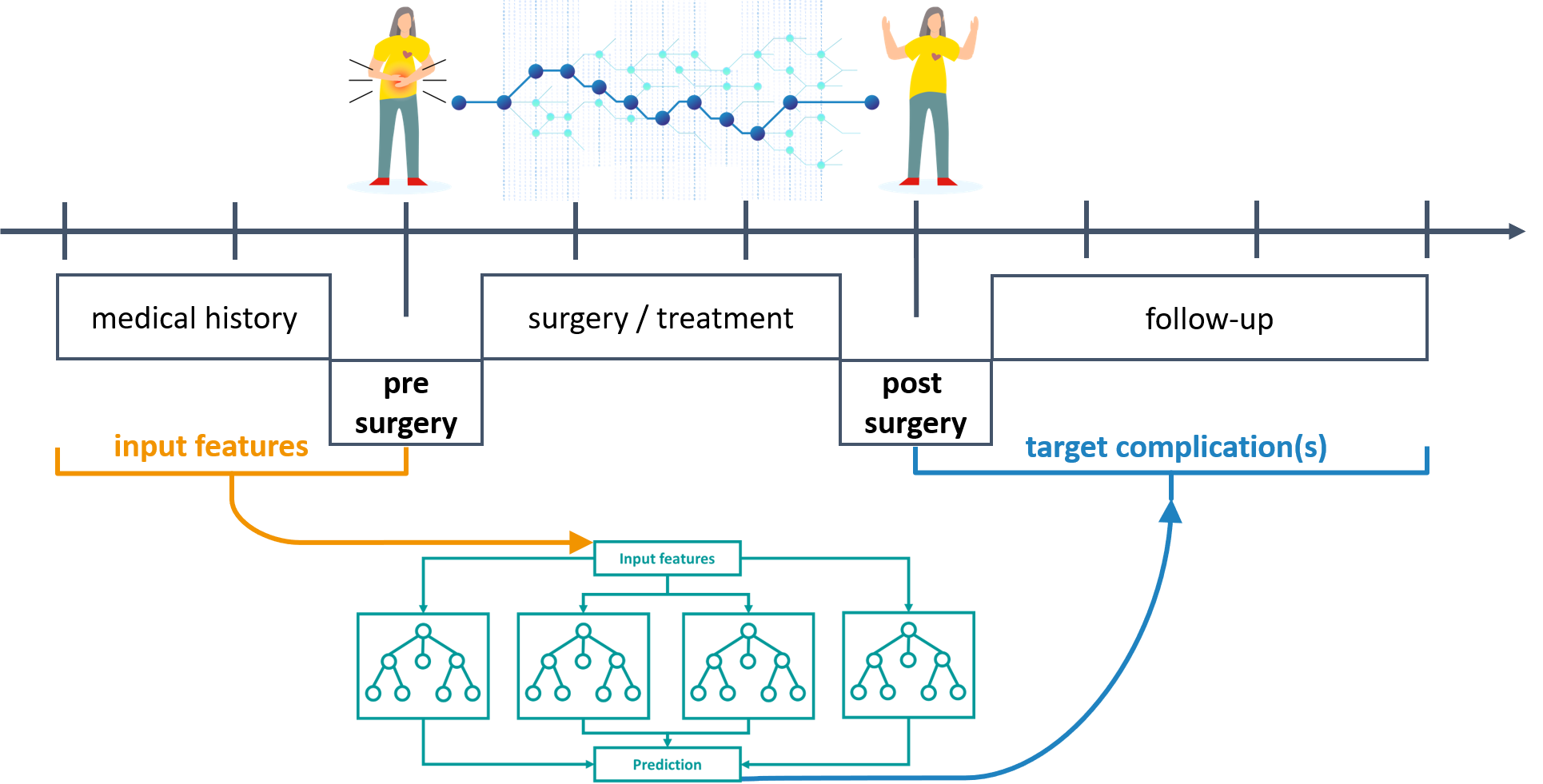
Example project: KIPeriOP
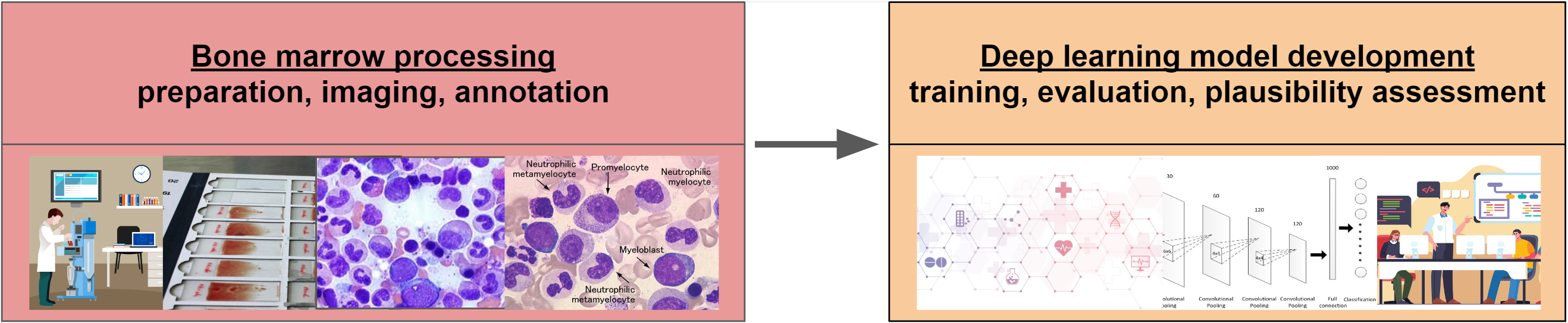
Example project: BMDeep
Example project: CTG
2126 fetal cardiotocograms (CTGs) were automatically processed and the respective diagnostic features measured. The CTGs were also classified by three expert obstetricians and a consensus classification label assigned to each of them. Classification was both with respect to a morphologic pattern (A, B, C. …) and to a fetal state (N, S, P). Therefore the dataset can be used either for 10-class or 3-class experiments
- The dataset consists of 2070 observations of 23 features.
- In the following, we consider the binary classification task {suspect, pathological} vs. normal
Example project: CTG
- “Cardiotocography (CTG) is a technique used to monitor the fetal heartbeat and uterine contractions during pregnancy and labour. The machine used to perform the monitoring is called a cardiotocograph.”
- “CTG monitoring is widely used to assess fetal well-being by identifying babies at risk of hypoxia (lack of oxygen). CTG is mainly used during labour.”
- Figure: “The display of a cardiotocograph. The fetal heartbeat is shown in orange, uterine contractions are shown in green, and the small green numbers (lower right) show the mother’s heartbeat.”

Task types in supervised learning
- Focus in this course: binary classification
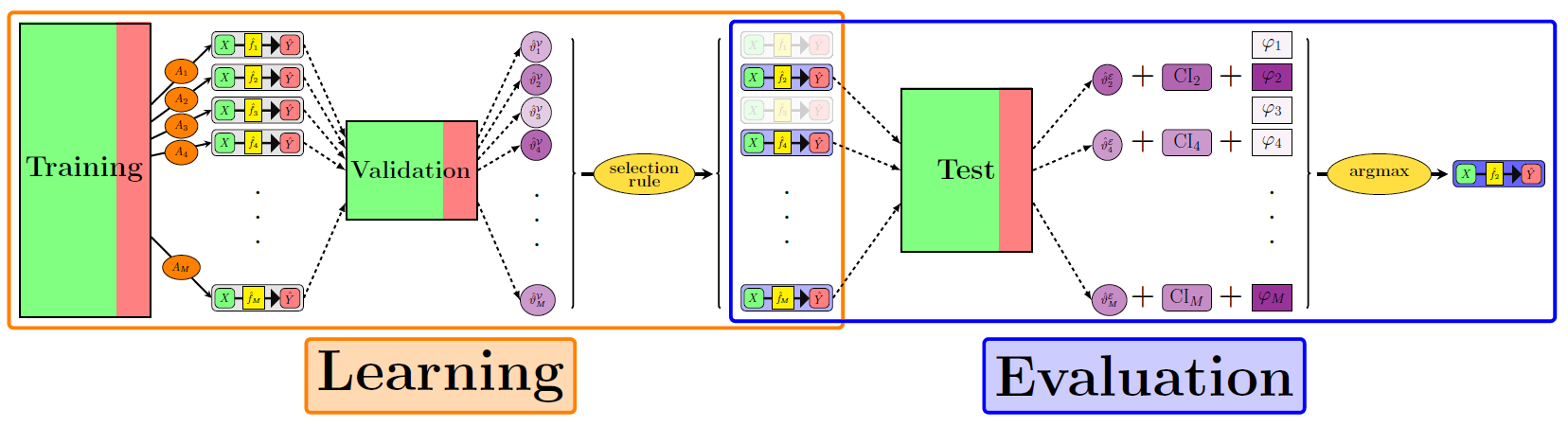
ML workflow
Types of evaluation studies
- This course is about evaluation studies assessing (some sort of) performance of prediction models or algorithms
- This course is not about assessing the downstream / real-world (clinical) utility
- Furthermore, we may distinguish between:
- internal validation: development and evaluation take place in different samples from the same population (in-distrubution)
- external validation: an independent evaluation (different time and/or reseach group)
- internal-external validation: during development, we try to assess the generalizability (transferability) to another context
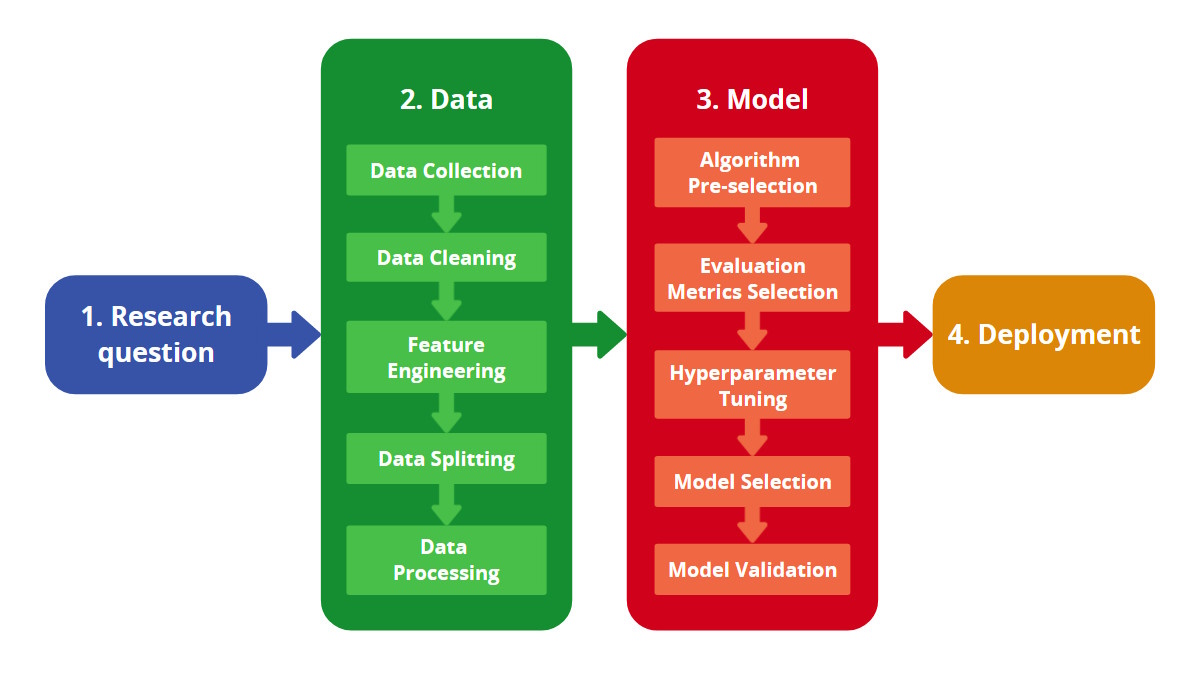
Applied ML: “standard” pipeline
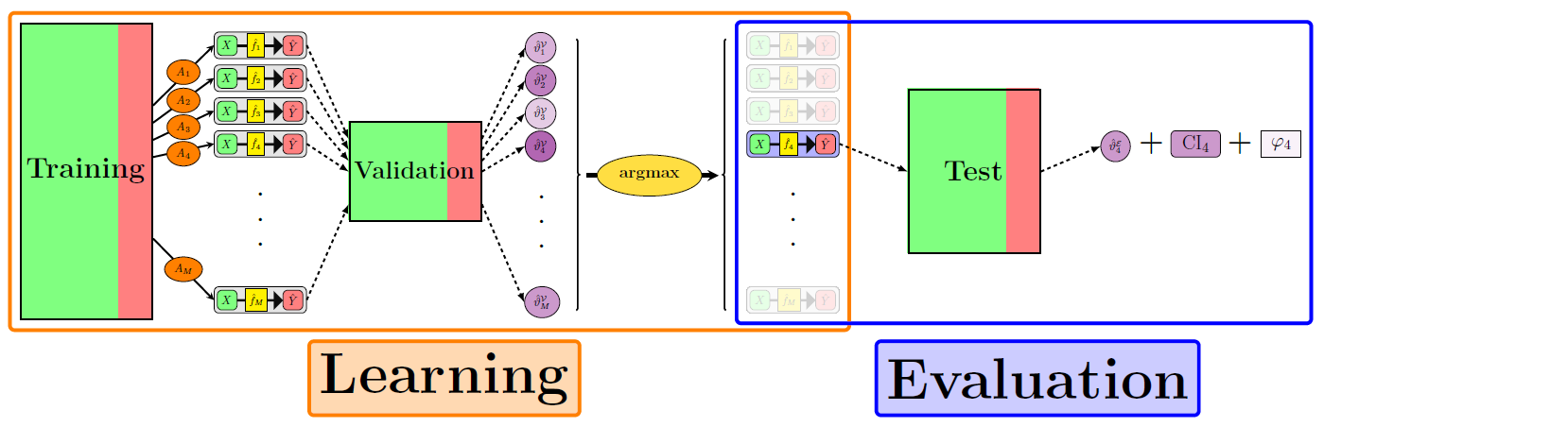
ML terminology is a mess
- Training, aka
- Development
- Derivation
- Learning
- Tuning, aka
- Validation
- Development
- Testing, aka
- Validation
- Evaluation
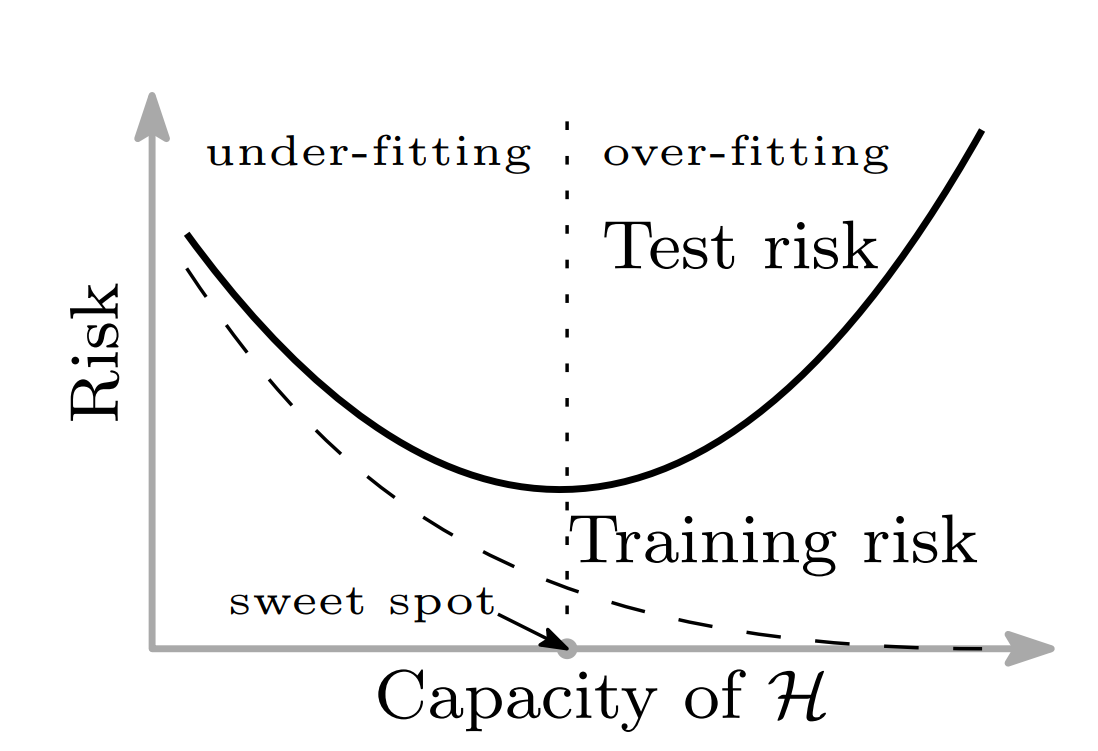
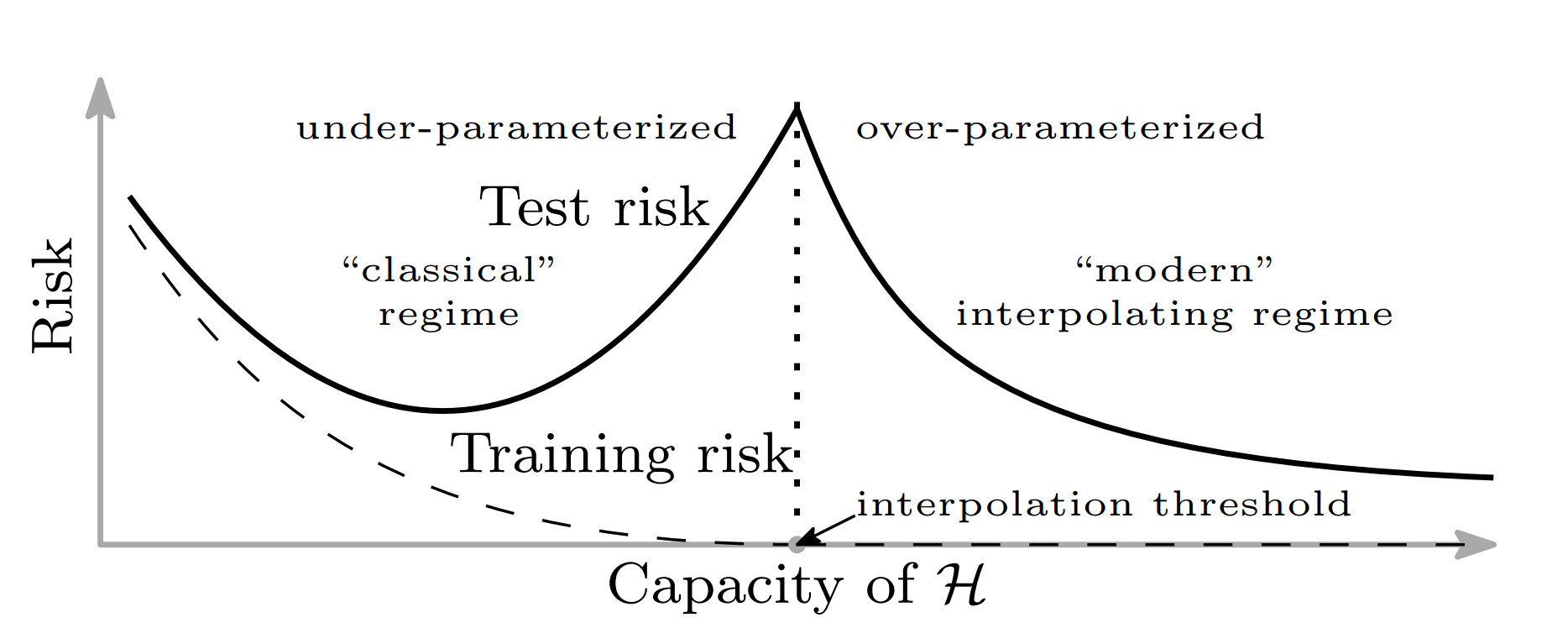
Bias variance trade-off
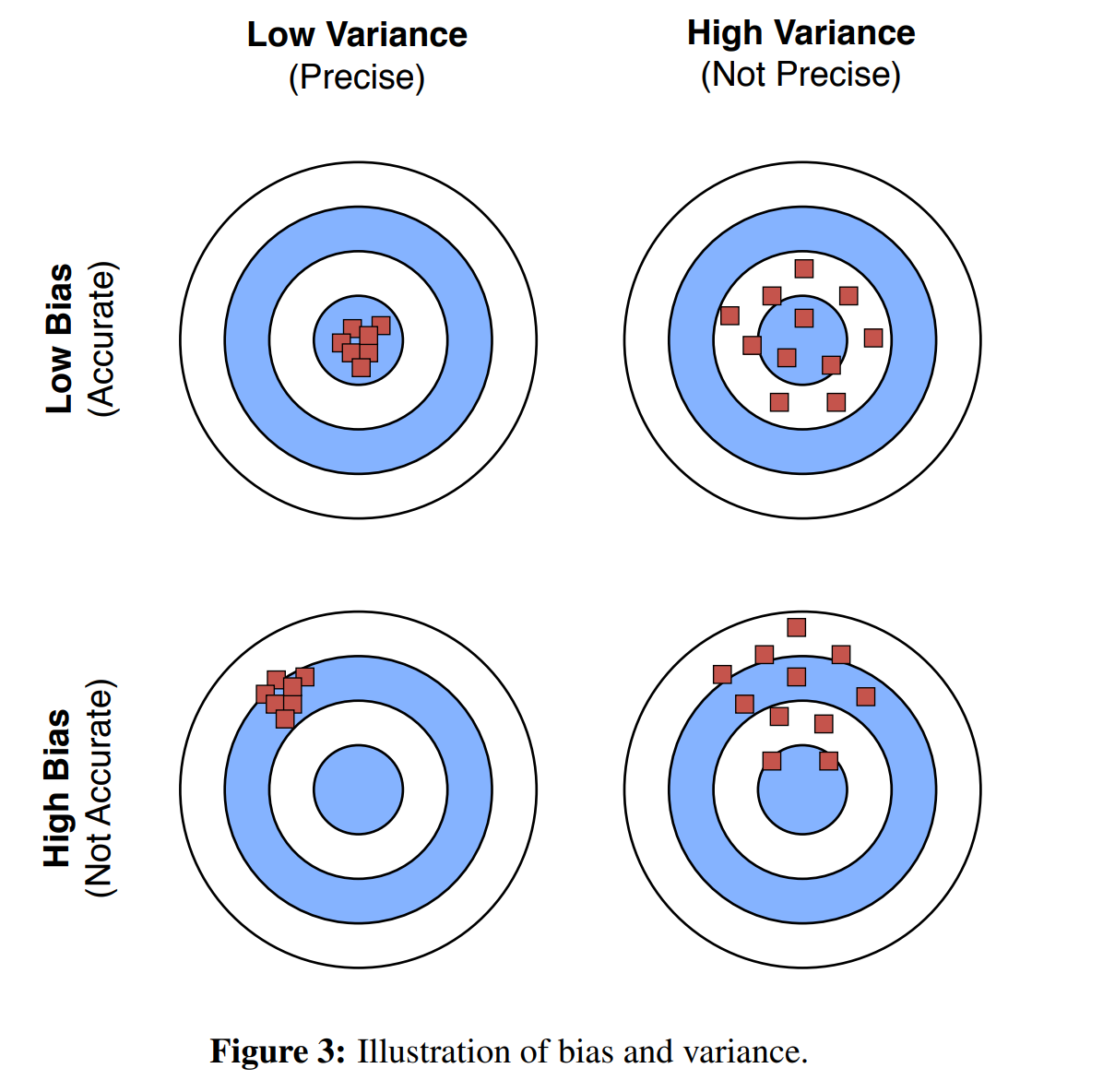
Bias variance trade-off in ML
Bias variance trade-off in ML (modern)
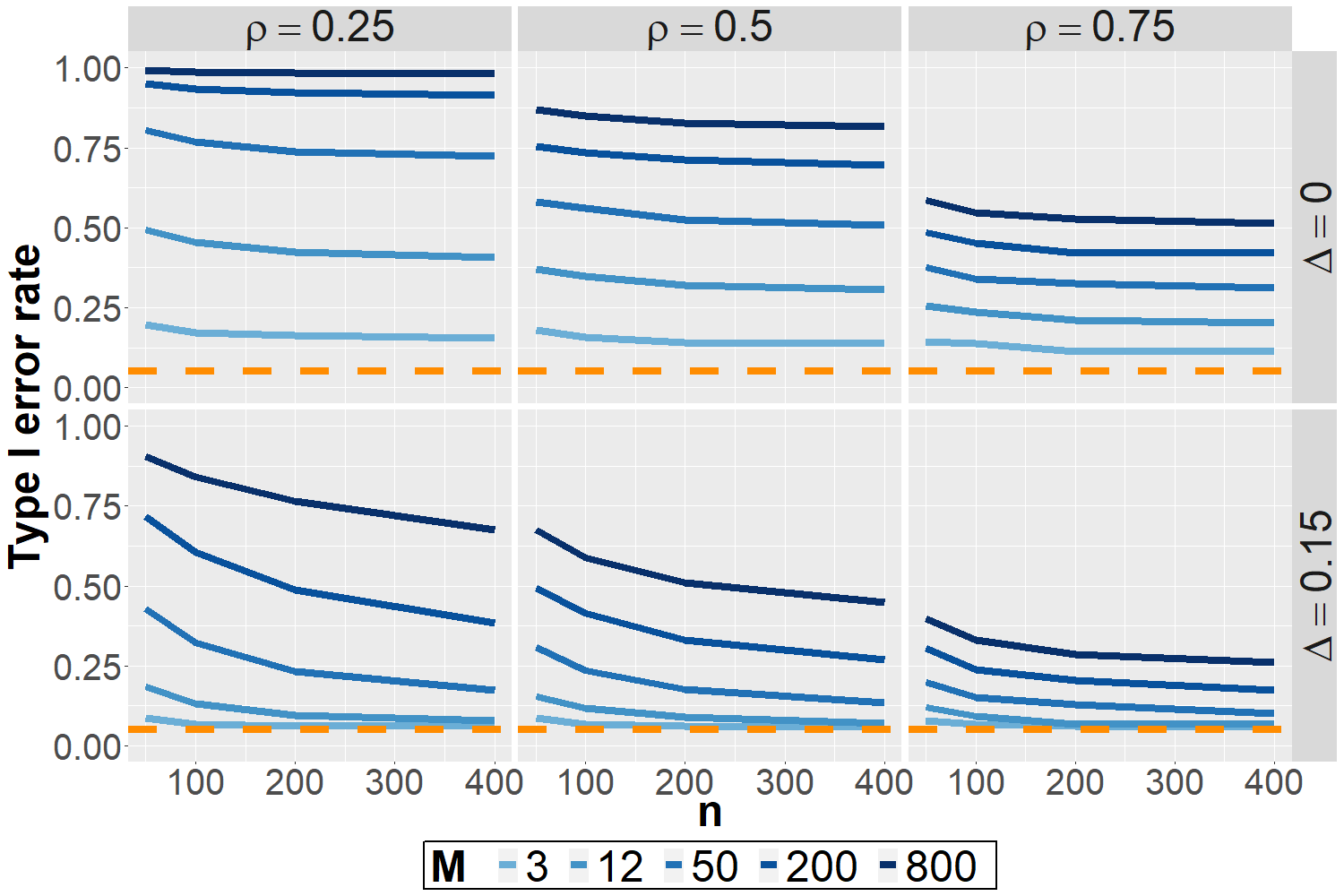
Selection-induced bias
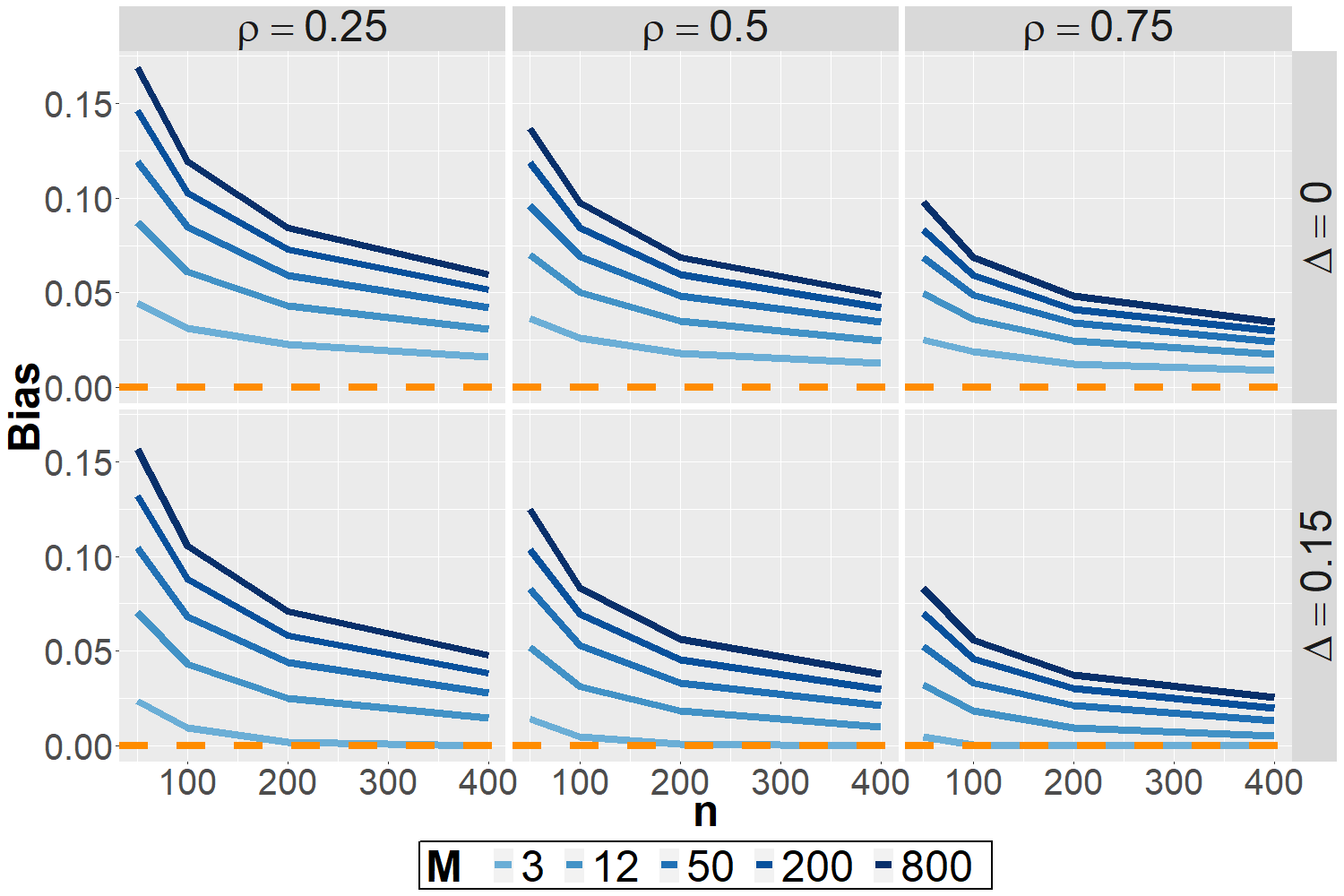
Selection-induced bias
No-free-lunch theorem
No single model (architecture) works best in all possible scenarios.
- Solutions
- extensive experiments
- inductive bias (prior knowledge)
- a mixture of both
Aleatoric and epistemic uncertainty in machine learning
- “Aleatoric (aka statistical) uncertainty refers to the notion of randomness, that is, the variability in the outcome of an experiment which is due to inherently random effects.”
- “Epistemic (aka systematic) uncertainty refers to uncertainty caused by a lack of knowledge, i.e., to the epistemic state of the agent.”
Typical ML evaluation pitfalls

Typical ML pitfalls

Interactive summary
Q1: What is NOT a typical pitfall in ML evaluation studies?
- data augmentation before data splitting
- ignoring temporal dependencies
- only evaluate each model once
- combined model tuning and evaluation on the test data
Q1: What is NOT a typical pitfall in ML evaluation studies?
- data augmentation before data splitting
- ignoring temporal dependencies
- only evaluate each model once
- combined model tuning and evaluation on the test data
Q2: Three datasets are needed in the standard ML pipeline to counter …
- overfitting and underfitting
- overfitting and selection-induced bias
- underfitting and selection-induced bias
- overfitting, underfitting and selection-induced bias
Q2: Three datasets are needed in the standard ML pipeline to counter …
- overfitting and underfitting
- overfitting and selection-induced bias
- underfitting and selection-induced bias
- overfitting, underfitting and selection-induced bias
Q3: What type of validation does NOT exist?
- external-internal
- internal-external
- external
- internal
Q3: What type of validation does NOT exist?
- external-internal
- internal-external
- external
- internal
Q4: What is NOT a valid ML task?
- multil-label classification
- time-to-event analysis
- binary classification
- multi-class regression
Q4: What is NOT a valid ML task?
- multil-label classification
- time-to-event analysis
- binary classification
- multi-class regression
Q5: When comparing multiple models on the same dataset, the severity of selection-induced bias…
- decreases with model similarity
- decreases with the number of models
- decreases with spread of true performances
- increases with spread of true performances
Q5: When comparing multiple models on the same dataset, the severity of selection-induced bias…
- decreases with model similarity
- decreases with the number of models
- decreases with spread of true performances
- increases with spread of true performances
Questions
Performance metrics
Performance dimensions
- Discrimintation: can the model distinguish between the target classes?
- Calibration: are probability prediction accurate?
- Fairness: is discrimination similar in important subgroups?
- Uncertainty quantification: how good is the coverage of prediction intervals?
- Explainability: usually assessed in user studies…
Metric choice
- A deliberate metric choice is very important for a successful ML project.
- a sensible metric supports/guides development
- an unreasonable metric hinders development
- What is an optimal solution worth, if it is optimal w.r.t. to a suboptimal metric?
- Finding an adequate metric can be time consuming
- should be done early on (before any developments)
- should be based on discussion with important stakeholders (e.g. potential users)
- “Standard” / “default” metrics in the field:
- usually a good idea to report as well (secondary)
- often not sufficient as primary / sole metric
- Multiple interesting metrics?
- Development usually facilitated if there is a clear decision rule how to rank models (e.g. primary metric)
Metrics: R packages
- Metrics: https://CRAN.R-project.org/package=Metrics (2018-07-09)
- ModelMetrics: https://CRAN.R-project.org/package=ModelMetrics ( 2020-03-17)
- metrica: https://CRAN.R-project.org/package=metrica (2023-04-14)
- SurvMetrics: https://CRAN.R-project.org/package=SurvMetrics (2022-09-03)
- MetricsWeighted: https://CRAN.R-project.org/package=Metrics (2023-06-05)
Binary classification: confusion matrix
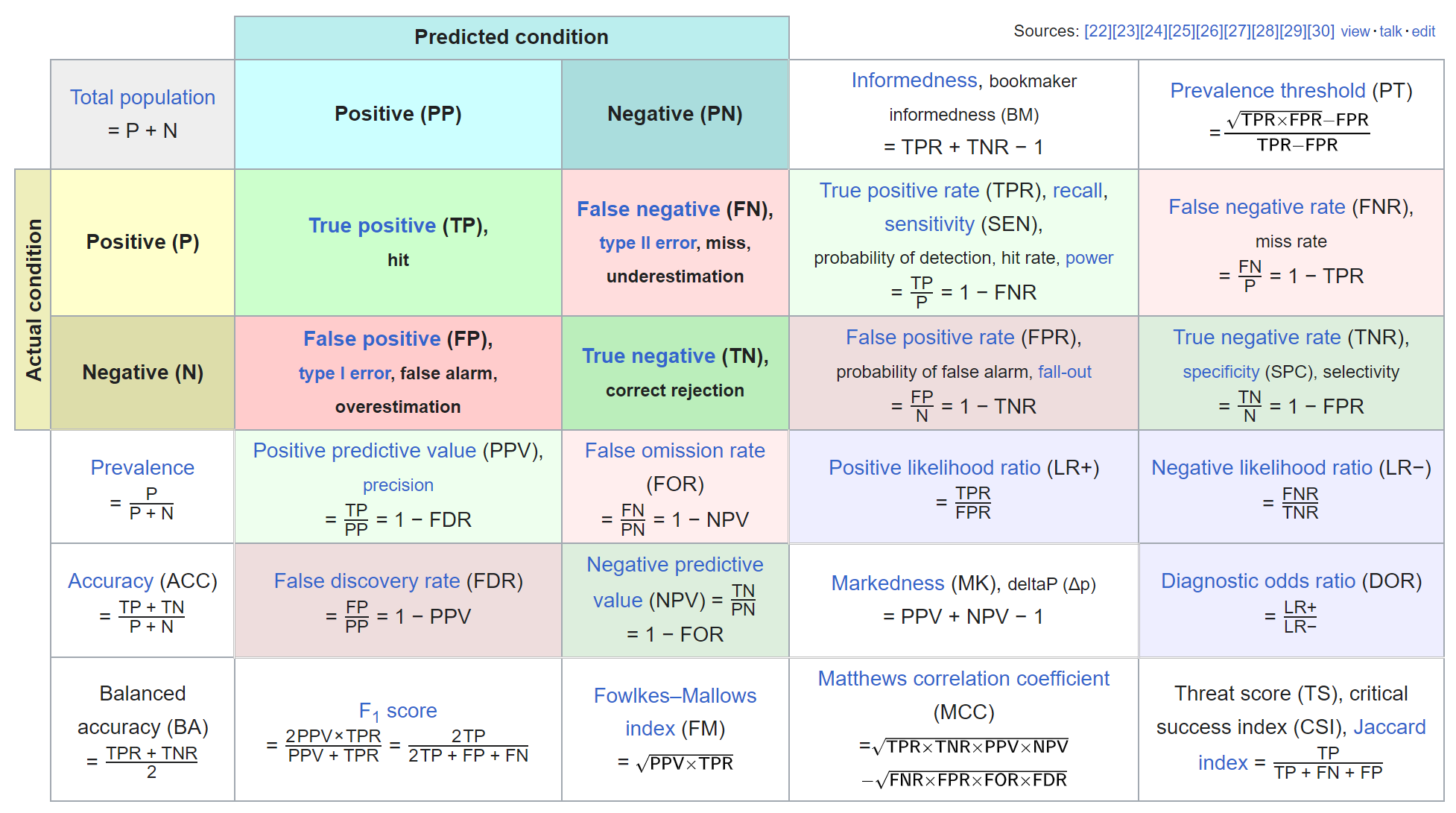
Evaluation data for CTG example (train-tune-test)
row_ids truth response prob.suspect prob.normal
1 3 normal normal 0.42001441 0.5799856
2 19 normal normal 0.00000000 1.0000000
3 28 normal normal 0.03574335 0.9642566
4 31 normal normal 0.04821049 0.9517895
5 47 normal normal 0.00000000 1.0000000
6 48 normal normal 0.00000000 1.0000000Binary classification: confusion matrix
caret::confusionMatrix(reference = data_eval_ttt_1$truth,
data = data_eval_ttt_1$response,
positive = "suspect")Confusion Matrix and Statistics
Reference
Prediction suspect normal
suspect 75 3
normal 15 319
Accuracy : 0.9563
95% CI : (0.9318, 0.9739)
No Information Rate : 0.7816
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.8656
Mcnemar's Test P-Value : 0.009522
Sensitivity : 0.8333
Specificity : 0.9907
Pos Pred Value : 0.9615
Neg Pred Value : 0.9551
Prevalence : 0.2184
Detection Rate : 0.1820
Detection Prevalence : 0.1893
Balanced Accuracy : 0.9120
'Positive' Class : suspect
Binary classification: discrimination metrics
- Accuracy: Probability of correct classification (actual \(=\) predicted)
- Sensitivity: Accuracy in positive class (cases, diseased, 1, TRUE)
- Specificity: Accuracy in negative class (controls, healthy, 0, FALSE)
- PPV: Accuracy in positive predictions
- NPV: Accuracy in negative predictions
- Balanced accuracy: Average of sensitivity and specificiy
Binary classification: weighted accuracy
a <- data_eval_ttt_1$truth == "suspect"
p <- data_eval_ttt_1$response == "suspect"
Metrics::accuracy(actual = a, predicted = p)[1] 0.9563107Binary classification: Risk prediction
- Binary classifier which not only predict the target variable (TRUE vs. FALSE), but also a probability of an event (TRUE), may also be called risk prediction models
- Often fundamental requirement in clinical predictive modelling
- As domain experts shall usually be supported (not replaced) by ML models / algorithms, a predicted risk can be much more informative and interpretable compared to a model only capable of predicting class labels
Receiver operating characteristic (ROC)
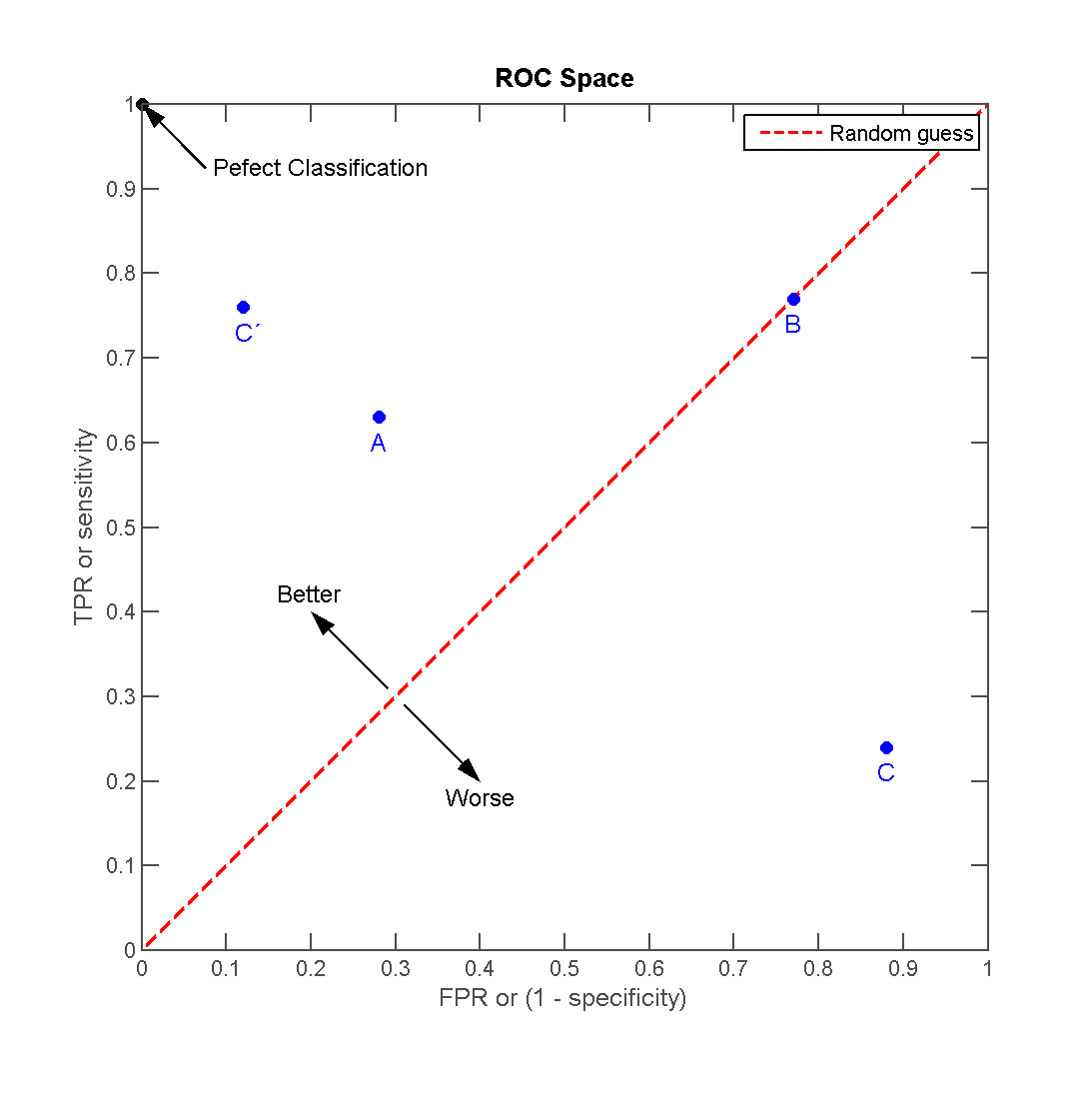
Receiver operating characteristic (ROC)
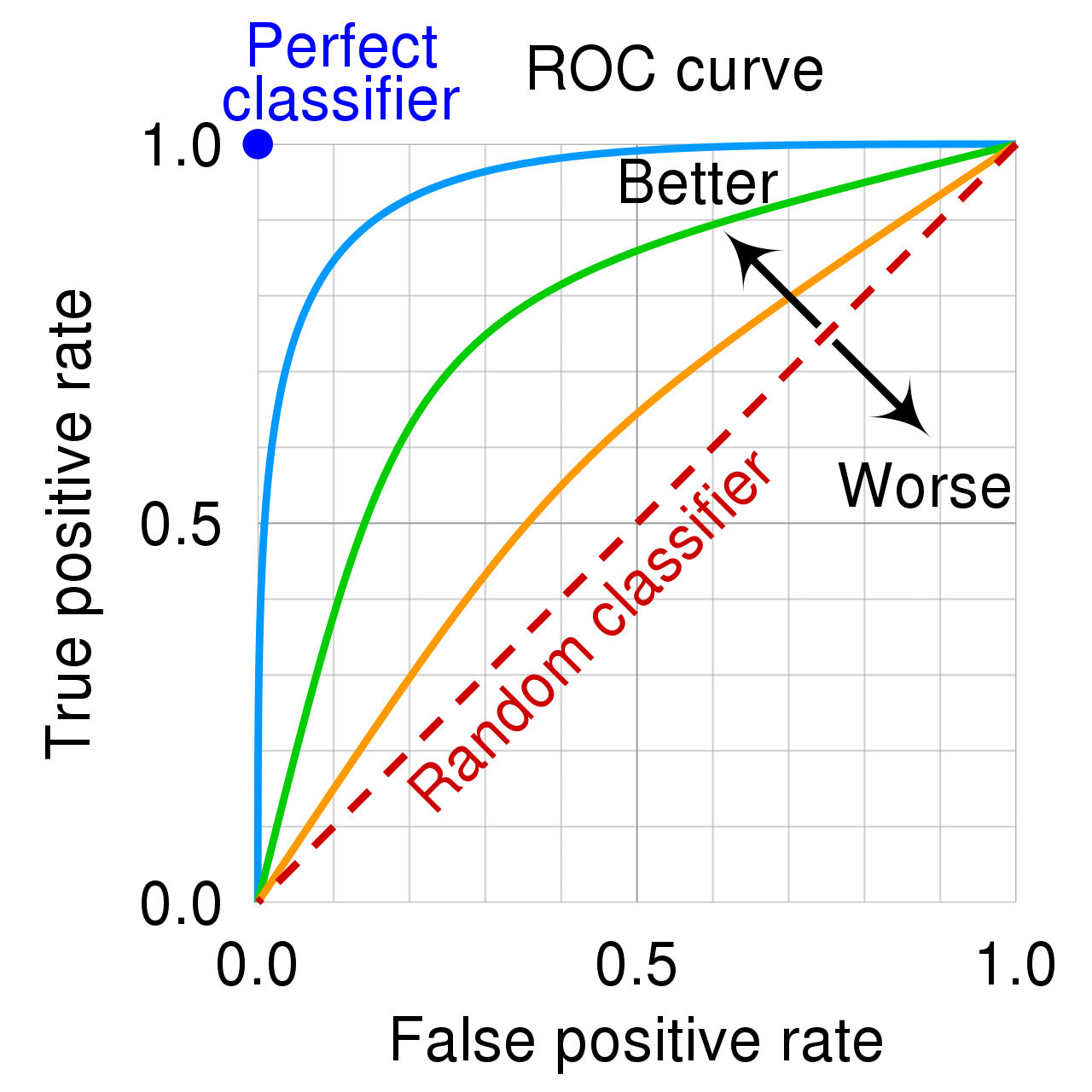
Receiver operating characteristic (ROC)
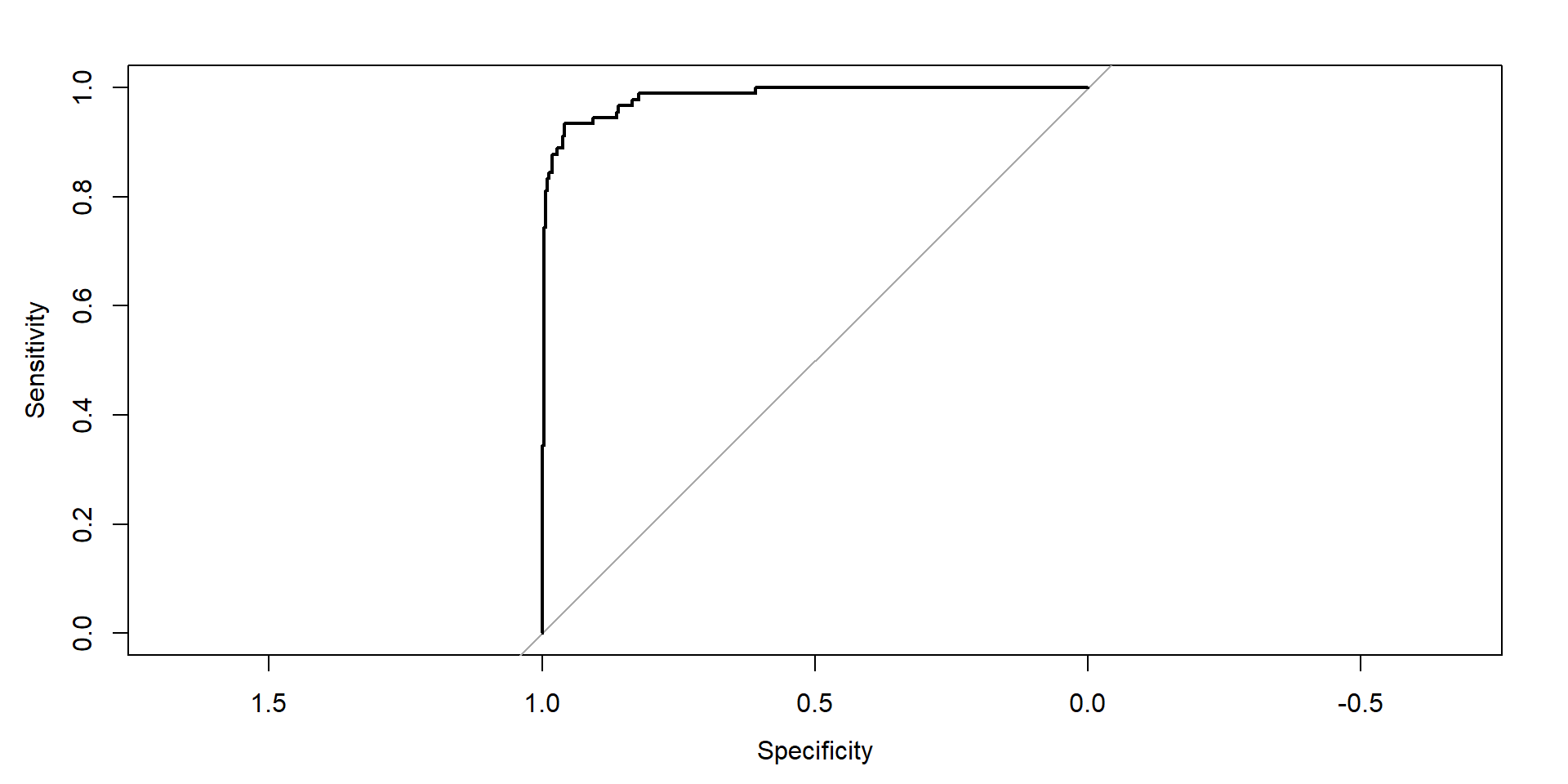
Binary classification: Area under the curve (AUC)
- Area under the (ROC) curve (also “c-statistic”, “concordance statistic”)
- Interpretation:
- Partial AUC: restrict specificity range
- Many different implementations (smoothed, with CI)
Binary classification: Area under the curve (AUC)
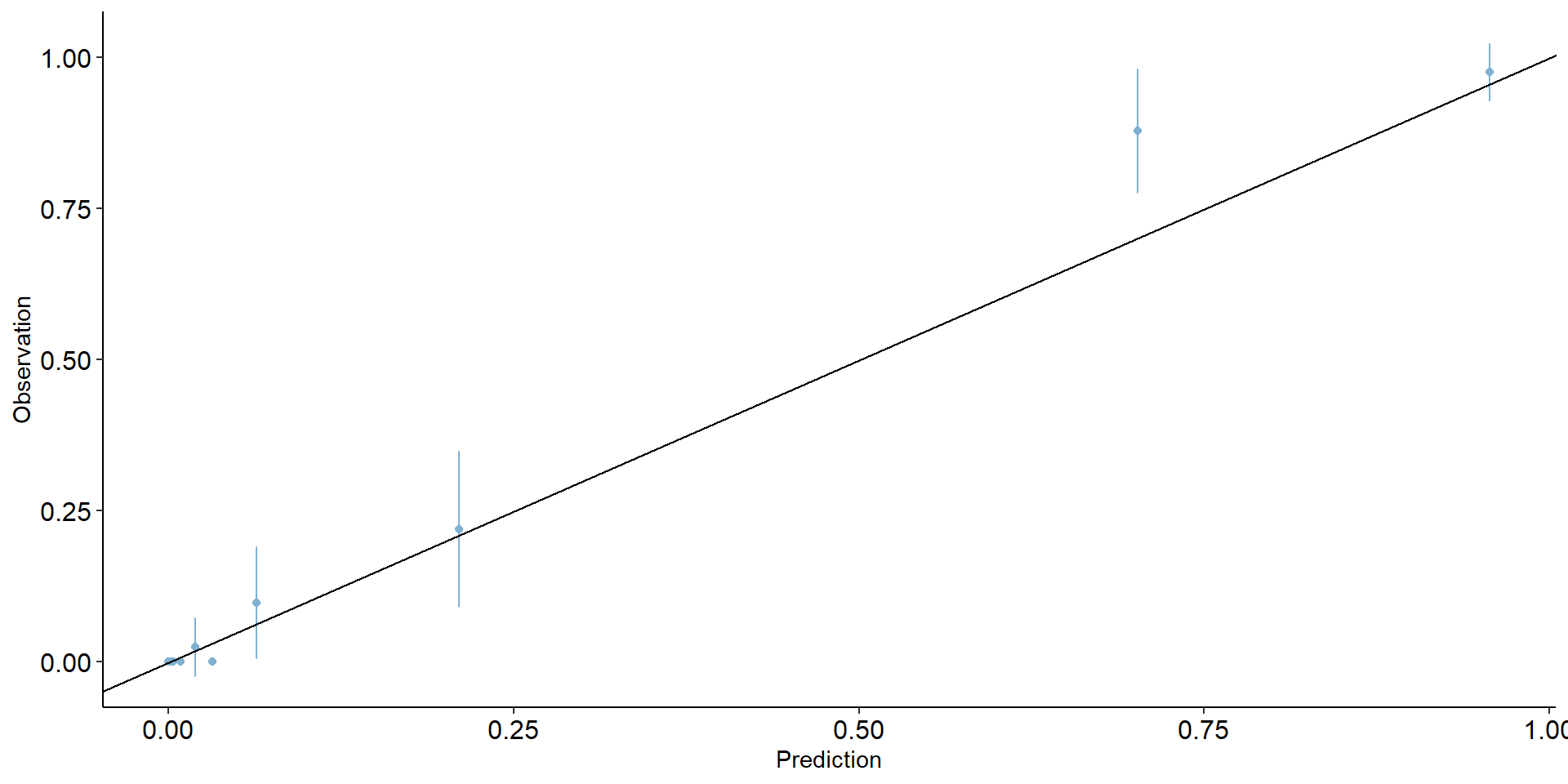
Binary classification: calibration plot
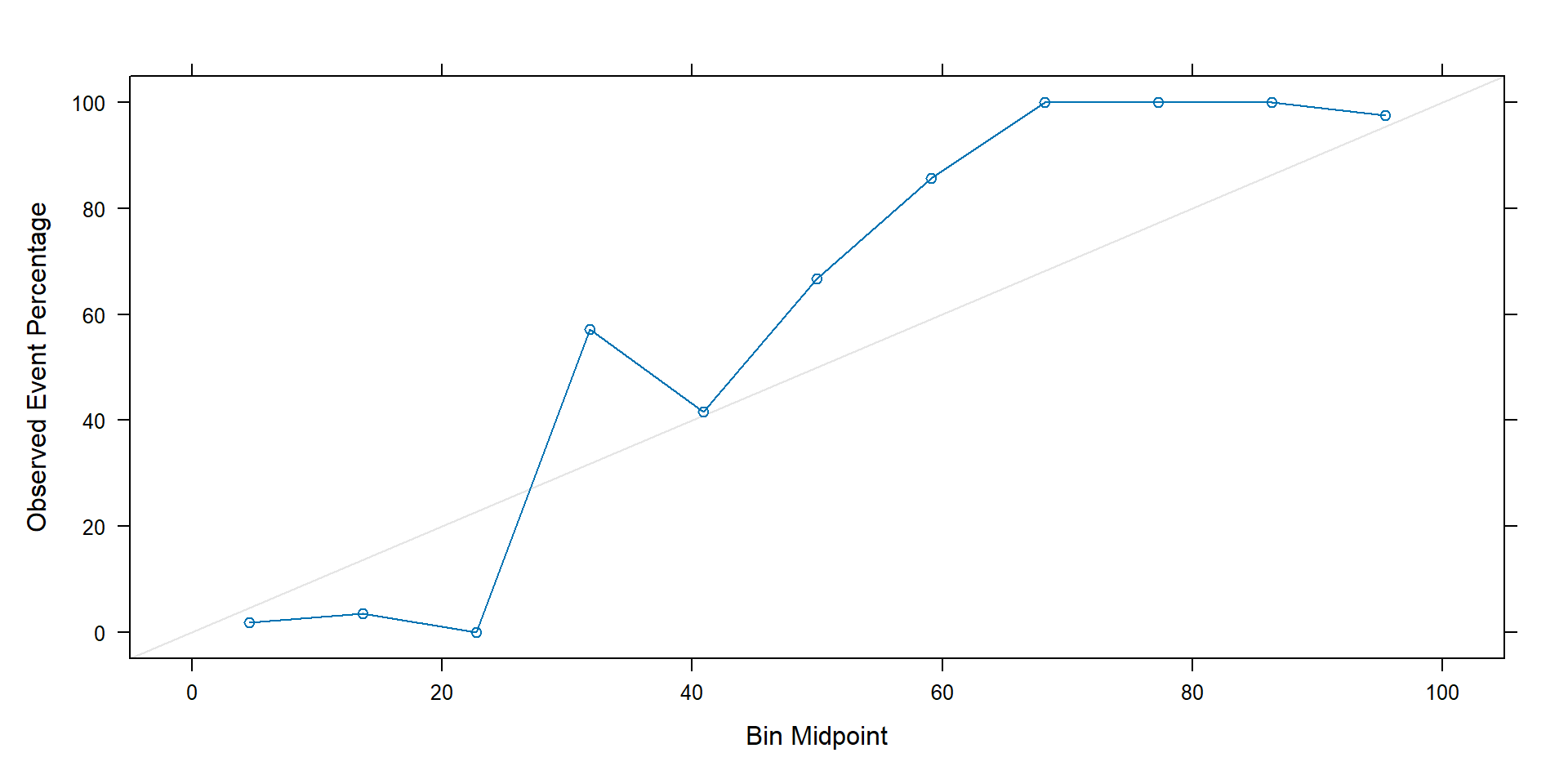
Binary classification: calibration plot
Binary classification: calibration plot
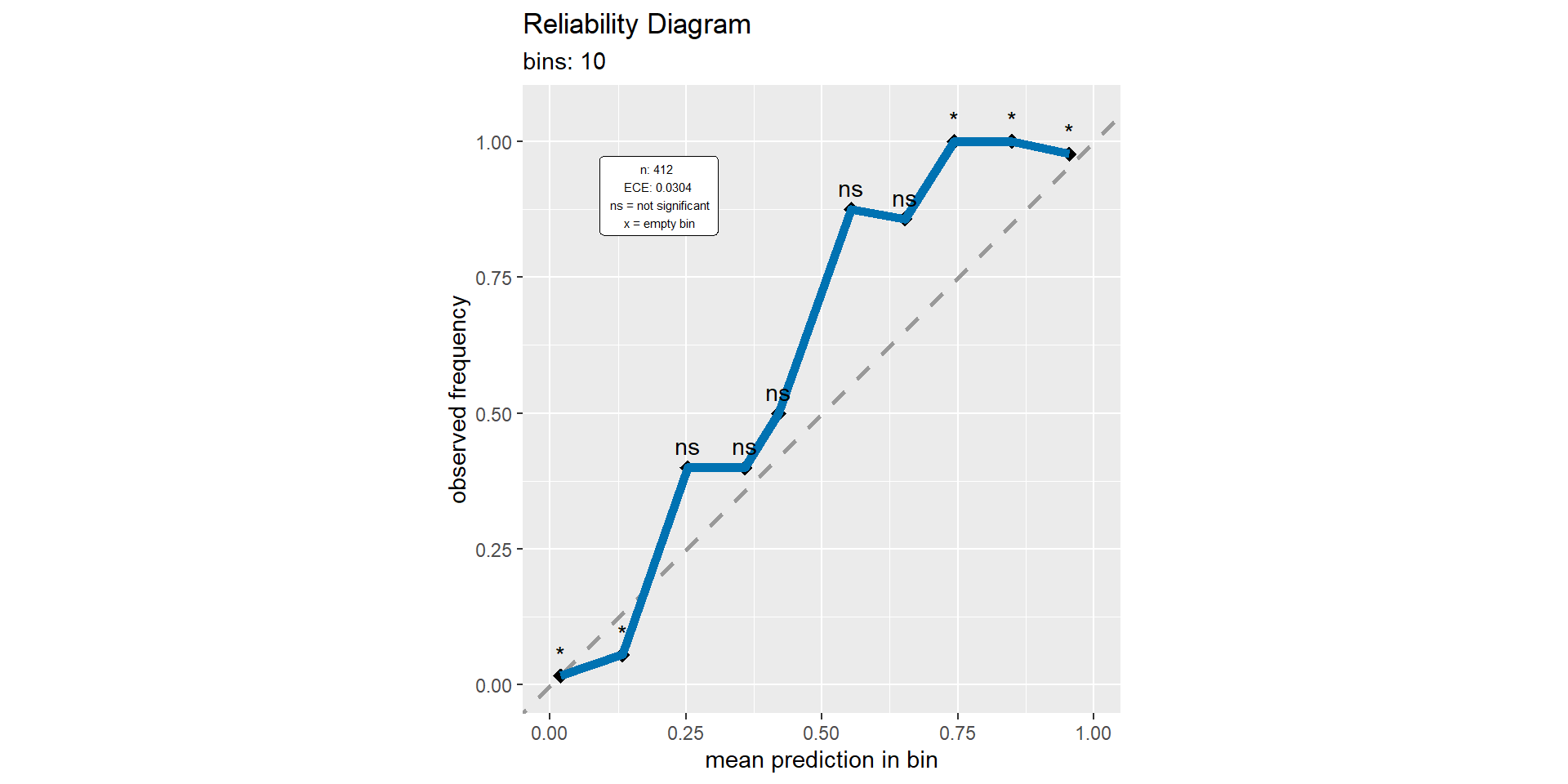
Binary classification: calibration metrics
- Expected calibration error (ECE)
- Maximum calibration error (MCE)
- Root mean squared error (from diagonal) (RSME)
- Calibration intercept (calibration-in-the-large)
- Calibration slope
Binary classification: calibration metrics
a <- data_eval_ttt_1$truth == "suspect"
p <- data_eval_ttt_1$prob.suspect
CalibratR::getECE(actual = a, predicted=p, n_bins=10)[1] 0.02858147Binary classification: calibration metrics
Calibration slope and intercept
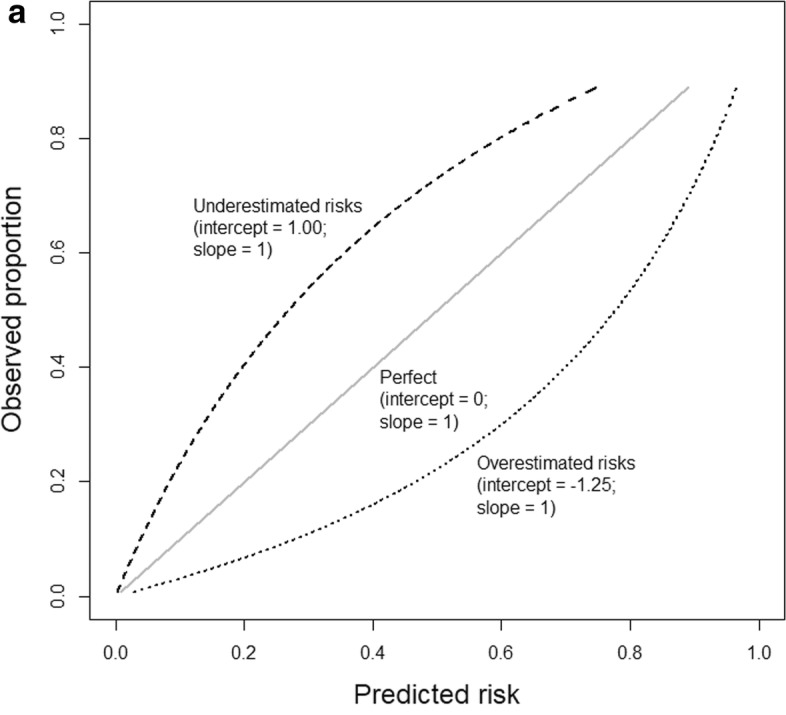
Calibration slope and intercept
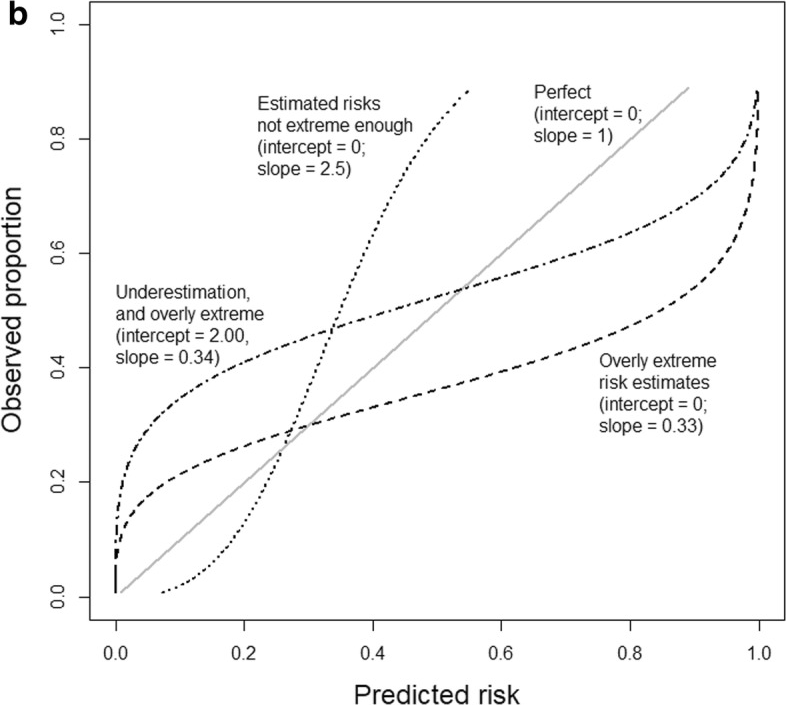
Binary classification: fairness - overview
- Predictive rate parity
- False positive rate parity
- False negative rate parity
- Accuracy parity
- Negative predictive value parity
- Specificity parity
- ROC AUC parity
- …
Multi-class classification: Confusion matrix
set.seed(123)
a <- rep(letters[1:5], times= (1:5)*20)
p <- sample(a, length(a))
caret::confusionMatrix(data = as.factor(p), reference=as.factor(a))Confusion Matrix and Statistics
Reference
Prediction a b c d e
a 2 4 2 5 7
b 1 7 9 11 12
c 4 9 14 15 18
d 5 6 20 22 27
e 8 14 15 27 36
Overall Statistics
Accuracy : 0.27
95% CI : (0.2206, 0.324)
No Information Rate : 0.3333
P-Value [Acc > NIR] : 0.9924
Kappa : 0.0338
Mcnemar's Test P-Value : 0.8813
Statistics by Class:
Class: a Class: b Class: c Class: d Class: e
Sensitivity 0.100000 0.17500 0.23333 0.27500 0.3600
Specificity 0.935714 0.87308 0.80833 0.73636 0.6800
Pos Pred Value 0.100000 0.17500 0.23333 0.27500 0.3600
Neg Pred Value 0.935714 0.87308 0.80833 0.73636 0.6800
Prevalence 0.066667 0.13333 0.20000 0.26667 0.3333
Detection Rate 0.006667 0.02333 0.04667 0.07333 0.1200
Detection Prevalence 0.066667 0.13333 0.20000 0.26667 0.3333
Balanced Accuracy 0.517857 0.52404 0.52083 0.50568 0.5200Multi-class classification: cost-sensitive metrics (BMDeep)
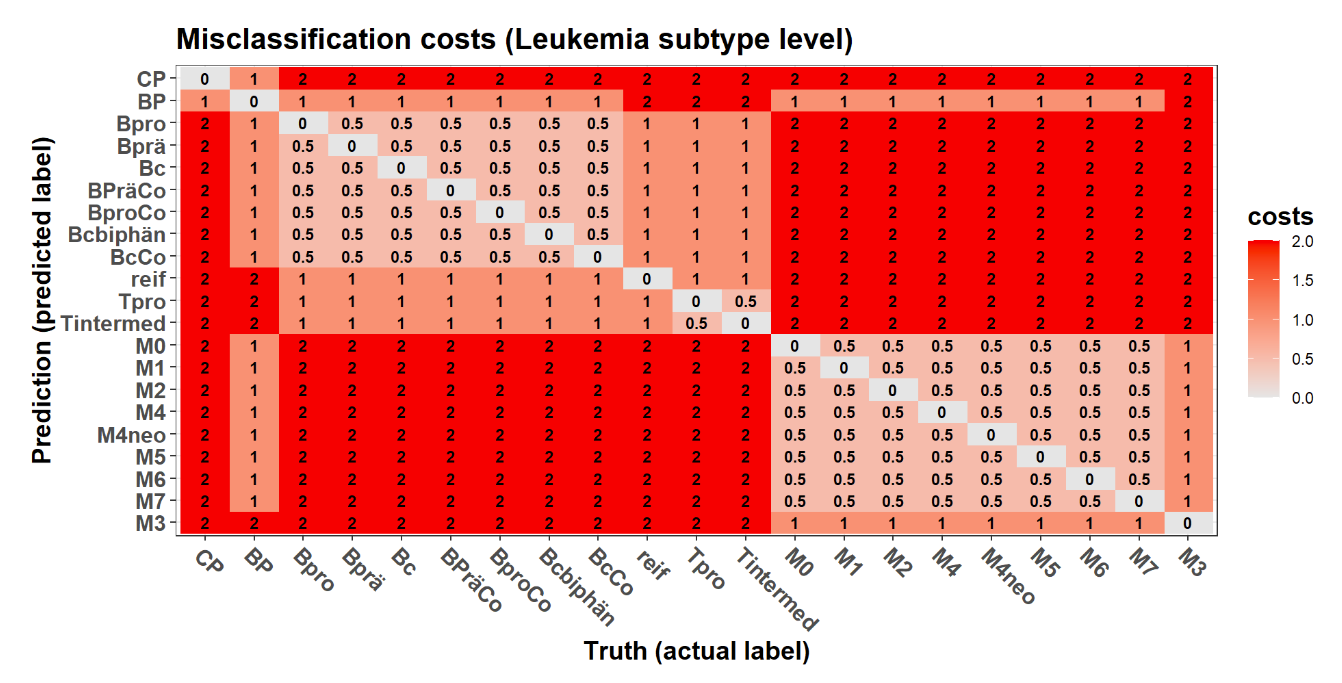
Multi-class classification: cost-sensitive metrics (BMDeep)
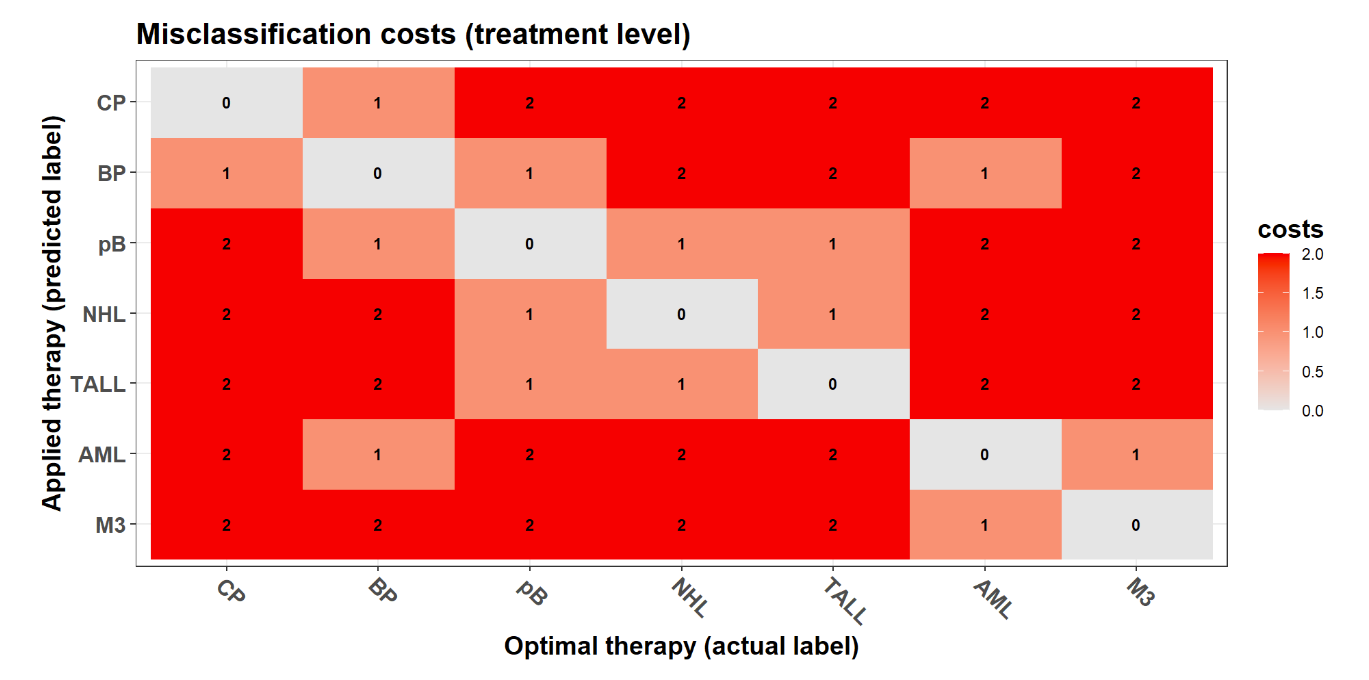
Regression: loss functions
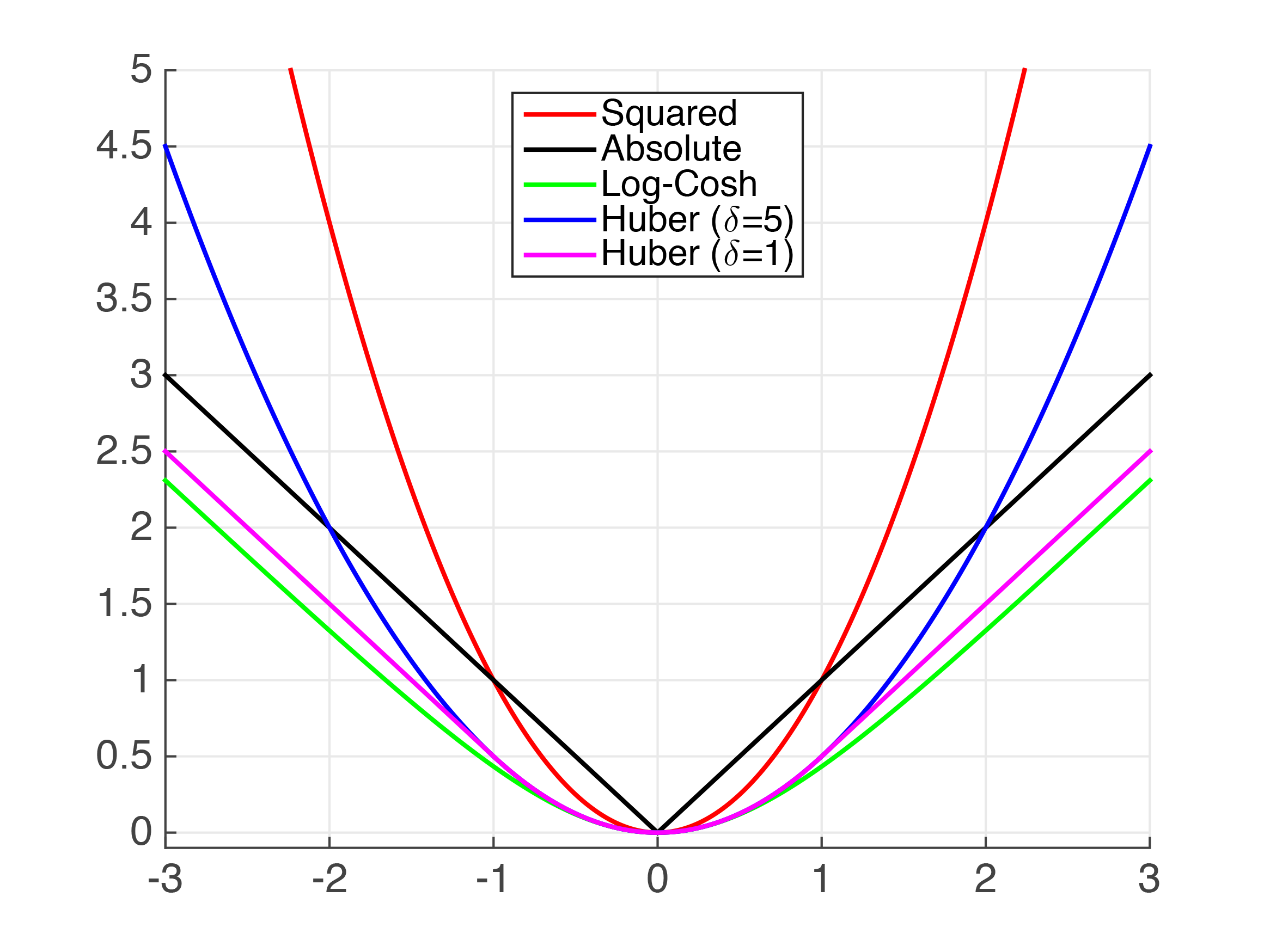
Regression: loss functions
- Symmetric loss functions used to define frequently used regression performance metrics, e.g.
- mean squared error
- mean absolute error
- For some regression tasks, custom performance metrics (based on asymmetric loss functions) may be more suitable
- for details, see references below
Survival analysis: overview
Mean absolute error (and other regression metrics)
- Not recommended as censoring is ignored
Concordance index/measure
- probability that of a randomly selected pair of patients , the patient with the shorter survival time has the higher predicted risk.
- most freuqently used
- several versions do exist
(Integrated) Brier score
Calibration slope
Metric choice
- A deliberate metric choice is very important for a successful ML project.
- a sensible metric supports/guides development
- an unreasonable metric hinders development
- What is an optimal solution worth, if it is optimal w.r.t. to a suboptimal metric?
- Finding an adequate metric can be time consuming
- should be done early on (before any developments)
- should be based on discussion with important stakeholders (e.g. potential users)
- “Standard” / “default” metrics in the field:
- usually a good idea to report as well (secondary)
- often not sufficient as primary / sole metric
- Multiple interesting metrics?
- Development usually facilitated if there is a clear decision rule how to rank models (e.g. primary metric)
Interactive summary
Q1: What is displayed in the confusion matrix?
- actual labels vs. predicted labels
- predicted labels vs. estimated probabilities
- actual labels vs. observed frequencies
- observed frequencies vs. estimated probabilities
Q1: What is displayed in the confusion matrix?
- actual labels vs. predicted labels
- predicted labels vs. estimated probabilities
- actual labels vs. observed frequencies
- observed frequencies vs. estimated probabilities
Q2: Which of the following metrics is NOT applicable to all binary classifiers?
- specificty
- accuracy
- positive predictive value
- area under the curve
Q2: Which of the following metrics is NOT applicable to all binary classifiers?
- specificty
- accuracy
- positive predictive value
- area under the curve
Q3: Calibration assesses …
- predicted probabilities relative to observed class labels
- predicted probabilities relative to observed class frequencies
- predicted class labels relative to the majority class
- predicted class labels relative to the minority class
Q3: Calibration assesses …
- predicted probabilities relative to observed class labels
- predicted probabilities relative to observed class frequencies
- predicted class labels relative to the majority class
- predicted class labels relative to the minority class
Q4: Which of the following statements is true?
- only sensitivty OR specifity should be considered, not both
- overall accuracy is independed of class prevalences
- balanced accuracy can be calculated from the confusing matrix
- balanced accuracy can be calculated from the confusion matrix
Q4: Which of the following statements is true?
- only sensitivty OR specifity should be considered, not both
- overall accuracy is independed of class prevalences
- balanced accuracy can be calculated from the confusing matrix
- balanced accuracy can be calculated from the confusion matrix
Q5: Which metrics can be used for the same ML task?
- balanced accuracy and mean squared error
- mean squared error and maximum calibration error
- area under the curve and maximum calibration error
- mean squared error and area under the curve
Q5: Which metrics can be used for the same ML task?
- balanced accuracy and mean squared error
- mean squared error and maximum calibration error
- area under the curve and maximum calibration error
- mean squared error and area under the curve
Questions
Break
Data splitting
Motivation: overfitting

Motivation: selection-induced bias

Overview of approaches
- Train-tune-test split
- Cross-validation (CV)
- Bootstrapping
- Stratified splitting
- Grouped (blocked) splitting
- Nested splitting (nested CV)
- Special variants (e.g. for time-series data)
The “default” train-tune-test split

The “default” train-tune-test split
- (Was used so far in the previous section)
- Simple to implement
- Low computational effort
- Simple statistical inference
- Target: conditional performance, either of…
- model trained on “train” dataset
- model trained on “train” & “tune” datasets combined
Cross-validation (+ independent test set)
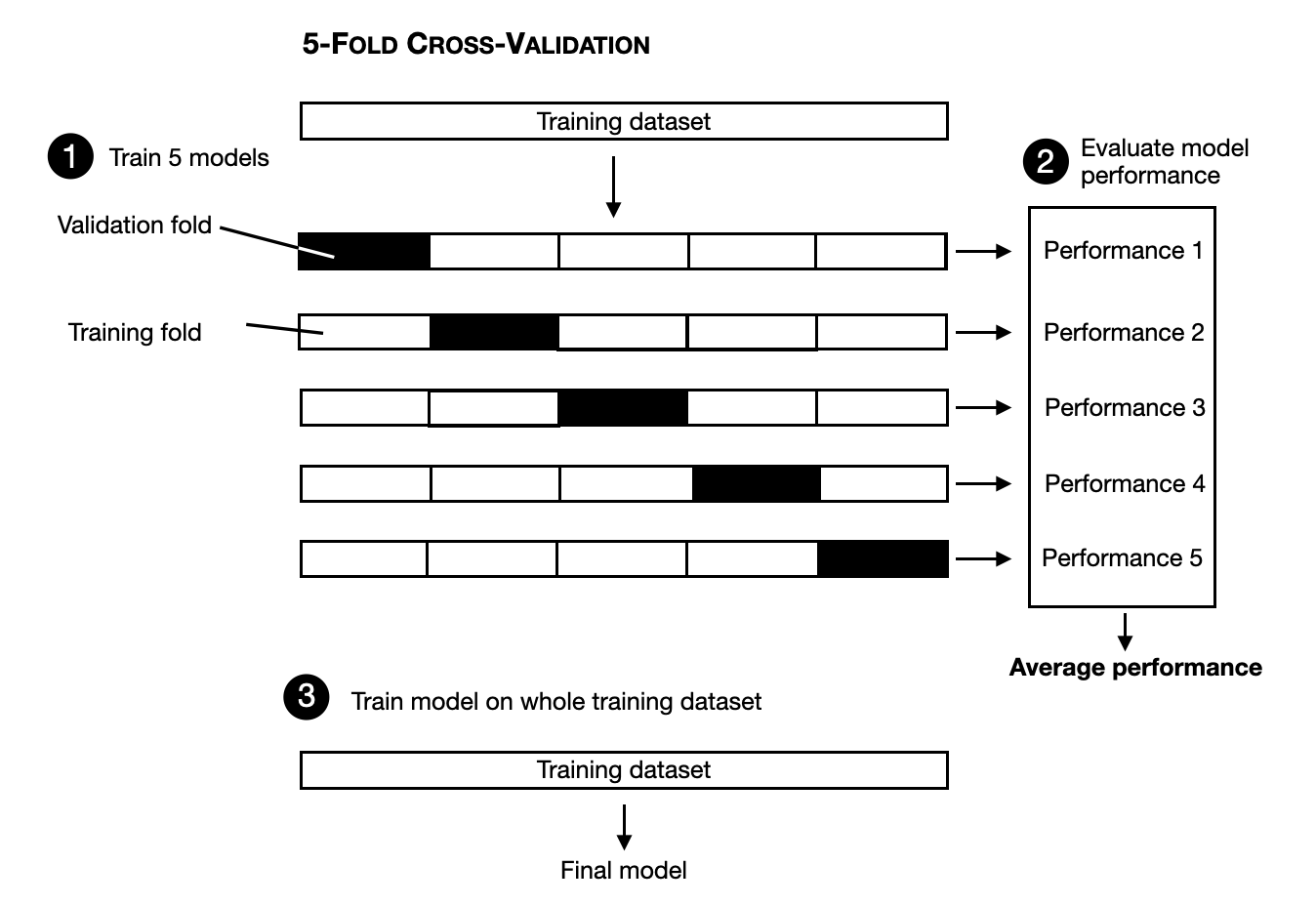
Cross-validation (+ independent test set)
- Cross-validation (cross-tuning):
- unconditional performance assessed
- increased computational burden (by number of folds \(K\))
- reduces danger of bad model selection for small samples
- Is the independent test set really required?
- It depends:
- Only model comparison needed: no
- Only performance assessment: no
- Both needed: yes, as (CV based) performance estimate of (CV) selected model can still be biased
- Solution: Nested cross-validation
Nested cross-validation
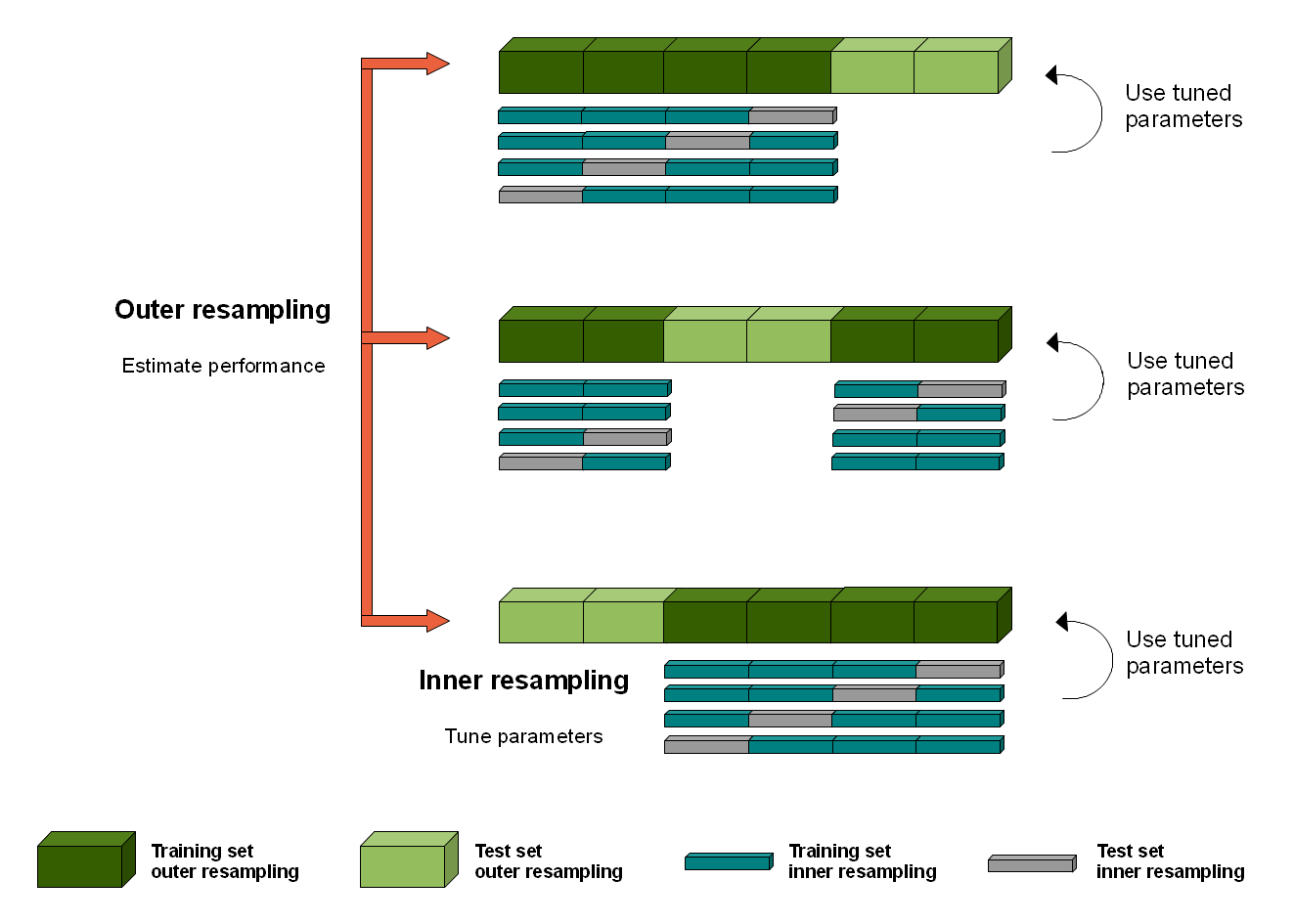
Computational complexity
- Train-tune-test: \(1\)
- K-fold CV: \(K\) (number of folds)
- Nested CV: \(K_{\text{inner}}\ \cdot K_{\text{outer}}\)
- Bootstrap: \(B\) (number of bootstrap repetitions)
- R-repeated K-fold CV: \(R \cdot K\)
- Leave-one-out CV: \(n_{\text{obs}}\) (number of observations)
General recommendations
- Trade off of between complexity (implementation, computation, analysis) and sample efficiency
- Large n: train/tune/test
- simple (implementation, computation, analysis)
- Small n: (nested) CV
- higher compute due to repeated sampling/training
- Optimal splitting (number of folds, ratio)
- no general accepted solution
- simulation…
- power calculation can guide minimal test set size
Splitting variants
- Stratification: should the distribution of certain variables be (approximately) equal in all datasets?
- In particular relevant for the outcome variable (classification)
- No/low costs, medium/high reward
- Grouping (also: blocking): Very relevant for hierarchical data
- Relevant question: What is the observational unit of your study?
- a (e.g. bone marrow) cell
- an image / a BM smear (= a collection of cells)
- a patient (= potentially multiple BM smears)
- If a lower hierarchy level is chosen, then splitting should be grouped by patient
There is more to it…
- Almost all data splitting techniques so far relied on random partitioning or sub-sampling
- For non-IID observations, e.g. time-series data, special data splitting methods are required (Schnaubelt, 2019)
- Question: What quantity are we really interested in?
- Observation: “generalization performance” is usually not defined very precisely in ML
Generalizability vs. transferability
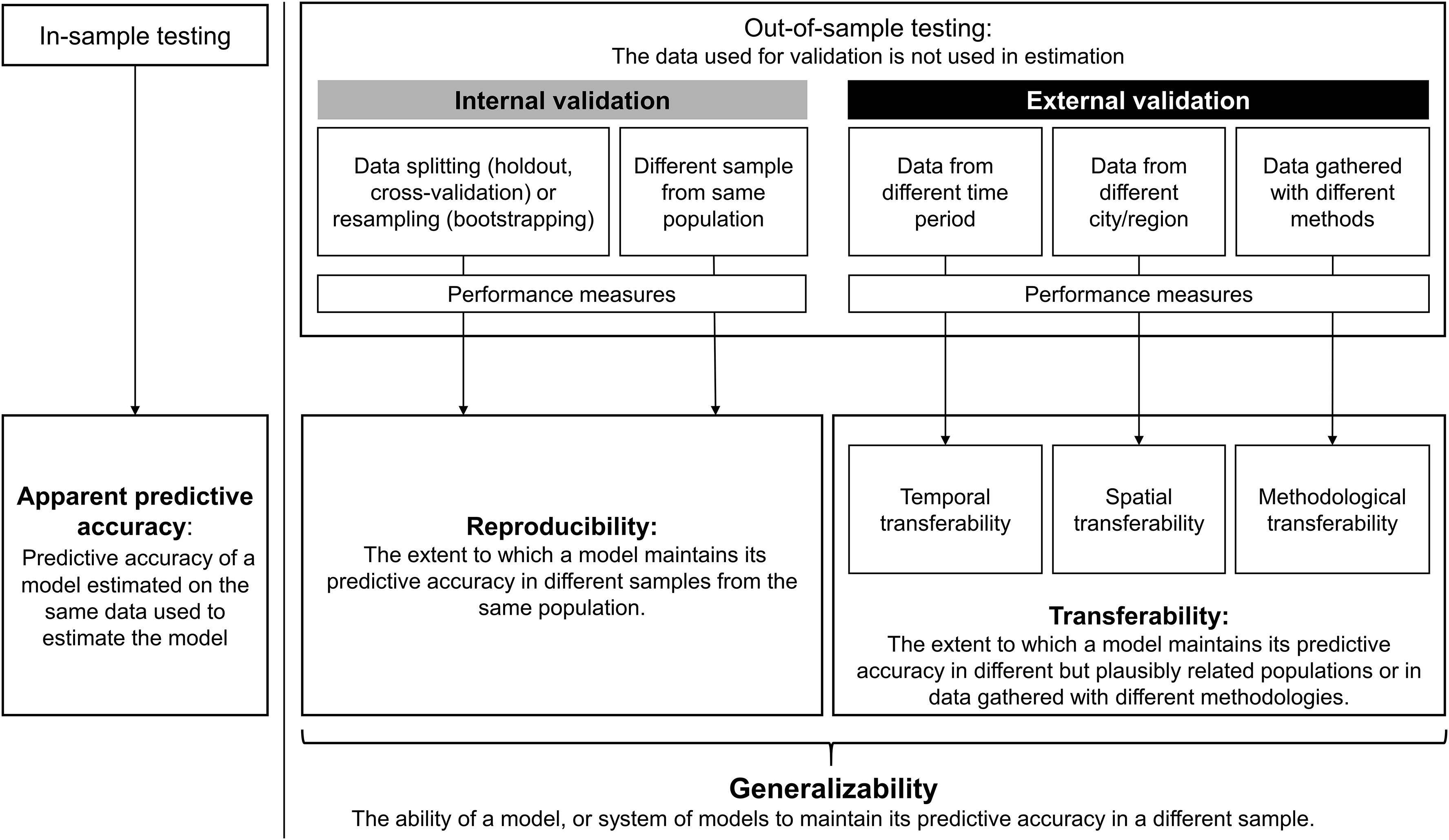
Estimand framework
Interactive summary
Q1: What is a commonly used data splitting technique?
- train-validation-test
- train-tune-test
- tune-validation
- cross-validation
Q1: What is a commonly used data splitting technique?
- train-validation-test
- train-tune-test
- tune-validation
- cross-validation
Q2: Which data splitting techniques are rarely used due to very high computational effort for model training?
- 5-fold cross-validation and the bootstrap
- the bootstrap and leave-one-out cross-validation
- 100-fold cross-validation and the train-tune-test split
- the bootstrap and the train-tune-test split
Q2: Which data splitting techniques are rarely used due to very high computational effort for model training?
- 5-fold cross-validation and the bootstrap
- the bootstrap and leave-one-out cross-validation
- 100-fold cross-validation and the train-tune-test split
- the bootstrap and the train-tune-test split
Q3: For large datasets, the classical “train-tune-test” split can be recommended due to…
- simple statistical analysis
- utilization of all observations for testing
- being the default method in many packages
- minimal computational effort
Q3: For large datasets, the classical “train-tune-test” split can be recommended due to…
- simple statistical analysis
- utilization of all observations for testing
- being the default method in many packages
- minimal computational effort
Q4: For small datasets, …
- a proper model comparison and evaluation is usually simpler
- nested data splitting techniques have increased relevance
- no data should be wasted for method evaluation
- the importance of power estimation is increased
Q4: For small datasets, …
- a proper model comparison and evaluation is usually simpler
- nested data splitting techniques have increased relevance
- no data should be wasted for method evaluation
- the importance of power estimation is increased
Q5: Which of the following statements is true?
- Stratification is required for hierarchical data structures
- Grouping is required for hierarchical data structures
- Stratification is required to retain the class distribution
- Grouping is required to retain the class distribution
Q5: Which of the following statements is true?
- Stratification is required for hierarchical data structures
- Grouping is required for hierarchical data structures
- Stratification is required to retain the class distribution
- Grouping is required to retain the class distribution
Questions
Statistical analysis
Goals of statistical inference
Estimation
Uncertainty quantification
- standard error
- confidence interval
Decision making
- hypothesis testing
Overview
Classical (frequentist) inference
Nonparametric methods
- bootstrap
- hierarchical bootstrap
Complex procedures
- mixed models
- Bayesian inference
Choice of comparator
- none (descriptive analysis)
- fixed performance threshold \(\vartheta_0 = 0.8\)
- another (established) prediction model: \(\vartheta_0 = \vartheta(\hat{f}_0)\)
- same test data
- paired comparison (!)
Evaluation data: train-tune-test (model comparison)
- Scenario: a random forest model was (hyperparameter) tuned on the train/tune data. The best/final model (w.r.t. tuning AUC) shall now be compared against an established elastic net model.
- Null hypothesis: \(\vartheta^\text{ranger}_* = \vartheta^\text{glmnet}_0\) (no difference in performance)
row_ids truth response_glmnet prob.suspect_glmnet prob.normal_glmnet
1: 3 normal normal 2.663412e-02 0.9733659
2: 19 normal normal 7.350806e-03 0.9926492
3: 28 normal normal 2.207659e-04 0.9997792
4: 31 normal normal 2.015005e-05 0.9999798
5: 47 normal normal 1.646076e-04 0.9998354
6: 48 normal normal 4.657763e-04 0.9995342
response_ranger prob.suspect_ranger prob.normal_ranger
1: normal 0.42001441 0.5799856
2: normal 0.00000000 1.0000000
3: normal 0.03574335 0.9642566
4: normal 0.04821049 0.9517895
5: normal 0.00000000 1.0000000
6: normal 0.00000000 1.0000000Data preparation
actual <- data_eval_ttt_2$truth
actual_01 <- (actual == "suspect") %>% as.numeric()
pred_glmnet <- data_eval_ttt_2$response_glmnet
pred_glmnet_01 <- (pred_glmnet== "suspect") %>% as.numeric()
correct_glmnet_01 <- (pred_glmnet_01 == actual_01) %>% as.numeric()
pred_ranger <- data_eval_ttt_2$response_ranger
pred_ranger_01 <- (pred_ranger== "suspect") %>% as.numeric()
correct_ranger_01 <- (pred_ranger_01 == actual_01) %>% as.numeric()Model 1: elastic net (glmnet)
Confusion Matrix and Statistics
Reference
Prediction suspect normal
suspect 69 8
normal 21 314
Accuracy : 0.9296
95% CI : (0.9005, 0.9524)
No Information Rate : 0.7816
P-Value [Acc > NIR] : 2.668e-16
Kappa : 0.7825
Mcnemar's Test P-Value : 0.02586
Sensitivity : 0.7667
Specificity : 0.9752
Pos Pred Value : 0.8961
Neg Pred Value : 0.9373
Prevalence : 0.2184
Detection Rate : 0.1675
Detection Prevalence : 0.1869
Balanced Accuracy : 0.8709
'Positive' Class : suspect
Model 2: random forest (ranger)
Confusion Matrix and Statistics
Reference
Prediction suspect normal
suspect 75 3
normal 15 319
Accuracy : 0.9563
95% CI : (0.9318, 0.9739)
No Information Rate : 0.7816
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.8656
Mcnemar's Test P-Value : 0.009522
Sensitivity : 0.8333
Specificity : 0.9907
Pos Pred Value : 0.9615
Neg Pred Value : 0.9551
Prevalence : 0.2184
Detection Rate : 0.1820
Detection Prevalence : 0.1893
Balanced Accuracy : 0.9120
'Positive' Class : suspect
Difference in proportions: t-test
Welch Two Sample t-test
data: correct_ranger_01 and correct_glmnet_01
t = 1.6531, df = 783.85, p-value = 0.09872
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.005005757 0.058403816
sample estimates:
mean of x mean of y
0.9563107 0.9296117 Difference in proportions: paired t-test
Paired t-test
data: correct_ranger_01 and correct_glmnet_01
t = 2.126, df = 411, p-value = 0.0341
alternative hypothesis: true mean difference is not equal to 0
95 percent confidence interval:
0.002012143 0.051385915
sample estimates:
mean difference
0.02669903 Difference in proportions: Newcombe interval
Nonparametric inference: Bootstrap
- Very versatile tool
- Minimal assumptions
- Not depending on asymptotics (large sample size)
Boostrap inference for single metric
Bootstrap inference for model comparison
delta_acc_paired <- function(correct_model_a, correct_model_b){
mean(correct_model_a - correct_model_b)
}
DescTools::BootCI(
x = correct_ranger_01,
y = correct_glmnet_01,
FUN = delta_acc_paired,
bci.method = "bca",
conf.level = 0.95
)delta_acc_paired lwr.ci upr.ci
0.026699029 0.002427184 0.049784828 Bootstrap inference for model comparison
delta_acc_paired_df <- function(df, i=1:nrow(df)){
df <- df[i,]
Metrics::accuracy(df$actual, df$pred_model_a) -
Metrics::accuracy(df$actual, df$pred_model_b)
}
df <- data.frame(actual = actual_01,
pred_model_a = pred_ranger_01,
pred_model_b = pred_glmnet_01)
boot::boot(data = df,
statistic = delta_acc_paired_df,
R = 1000) %>%
boot::boot.ci(conv=0.95, type="bca")BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
Based on 1000 bootstrap replicates
CALL :
boot::boot.ci(boot.out = ., type = "bca", conv = 0.95)
Intervals :
Level BCa
95% ( 0.0000, 0.0494 )
Calculations and Intervals on Original ScaleCI for difference in AUCs
rocc_glmnet <- pROC::roc(data_eval_ttt_2, response="truth", predictor="prob.suspect_glmnet", levels=c("normal", "suspect"))
rocc_ranger <- pROC::roc(data_eval_ttt_2, response="truth", predictor="prob.suspect_ranger", levels=c("normal", "suspect"))
plot(rocc_glmnet, col="blue")
plot(rocc_ranger, add=TRUE, col="orange")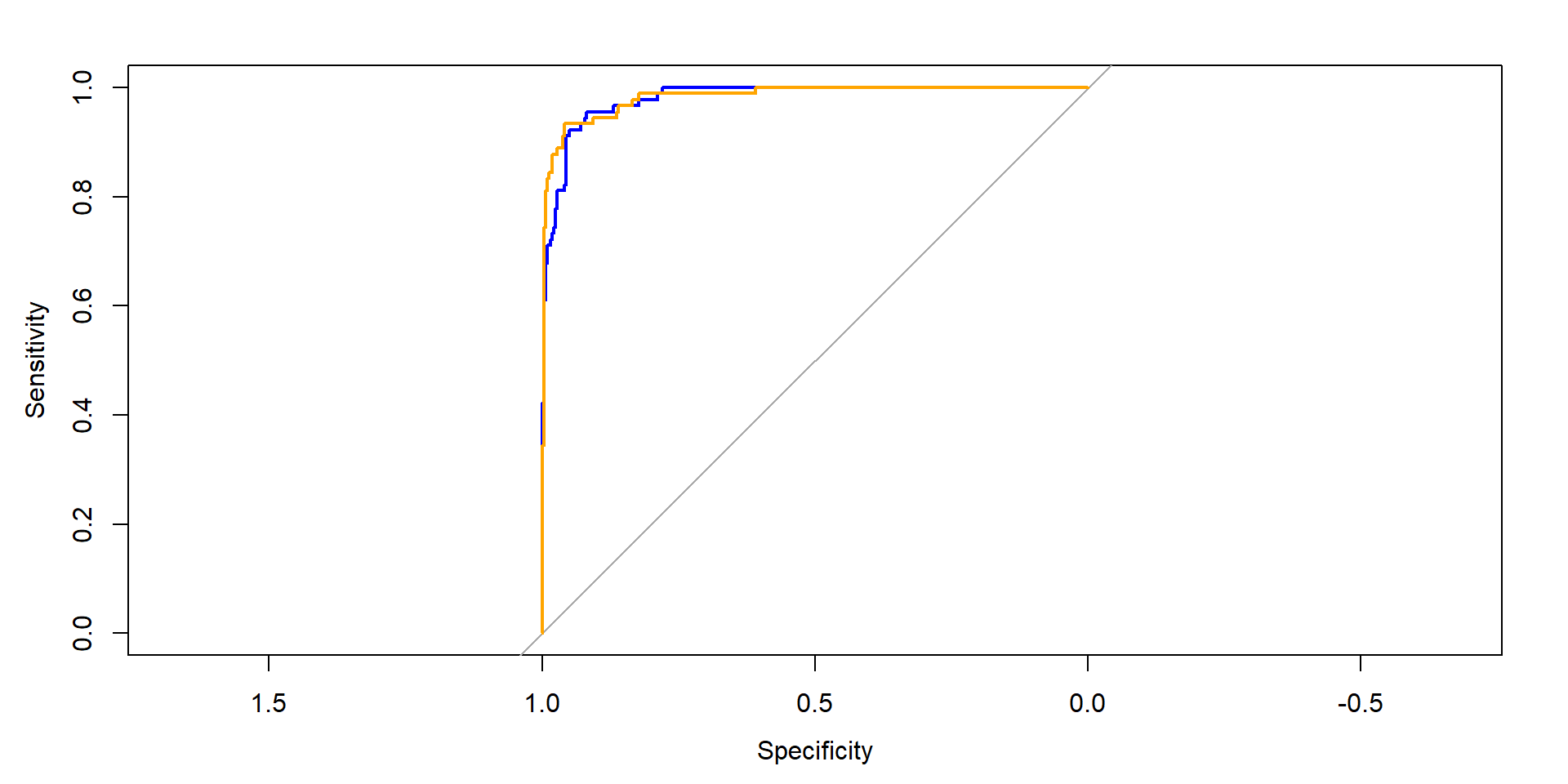
CI for difference in AUCs (Delong)
DeLong's test for two correlated ROC curves
data: rocc_ranger and rocc_glmnet
Z = 0.40273, p-value = 0.6871
alternative hypothesis: true difference in AUC is not equal to 0
95 percent confidence interval:
-0.009339915 0.014170832
sample estimates:
AUC of roc1 AUC of roc2
0.9830918 0.9806763 CI for difference in AUCs (bootstrap)
Bootstrap test for two correlated ROC curves
data: rocc_ranger and rocc_glmnet
D = 0.39739, boot.n = 2000, boot.stratified = 1, p-value = 0.6911
alternative hypothesis: true difference in AUC is not equal to 0
sample estimates:
AUC of roc1 AUC of roc2
0.9830918 0.9806763 UQ for (nested) CV
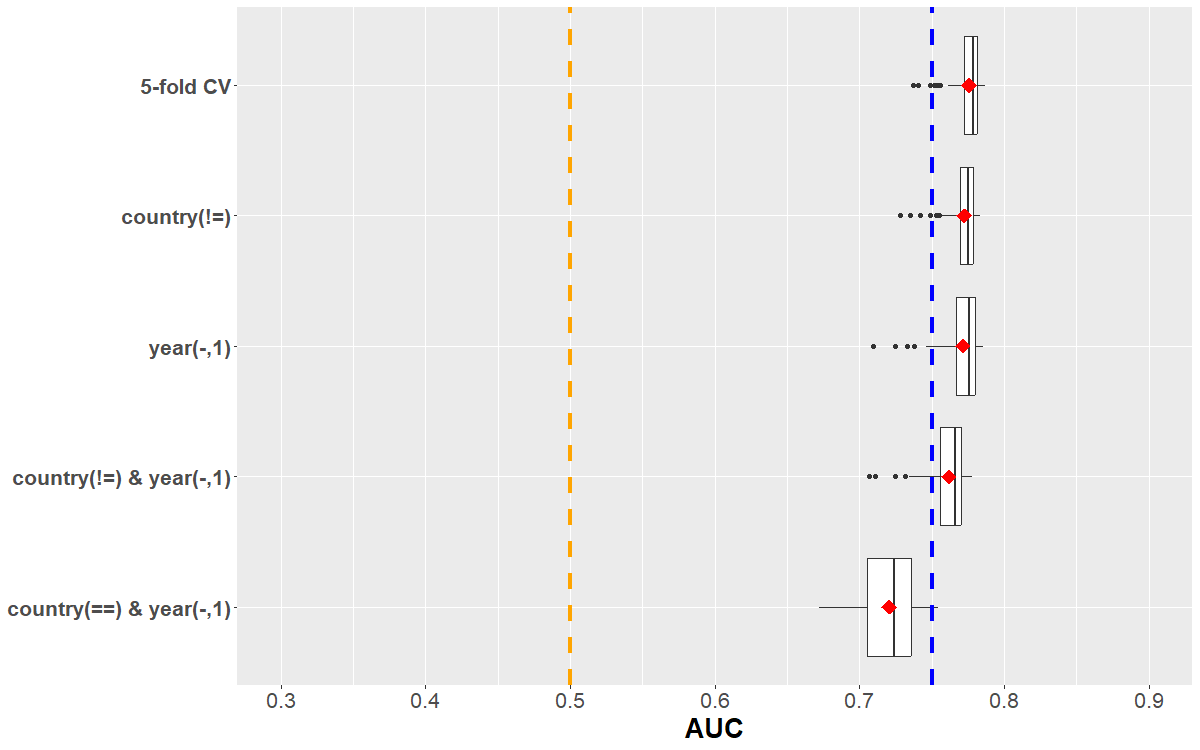
- Scenario: Algorithm evaluation or comparison in the outer loop of nested CV
- glmnet, ranger were tuned in the inner loop
- comparison of best hyperparameter combination of each approach
- on each fold (1:5), compare model that was trained on remaining observations
- How can we quantify uncertainty for (difference of) performance estimate?
- A few recommendations do exist (Raschka, 2018)
- We will focus again on the bootstrap and a hierarchical bootstrap variant
UQ for (nested) CV
fold row_ids truth response_glmnet prob.suspect_glmnet prob.normal_glmnet
1: 1 3 normal normal 3.057173e-02 0.9694283
2: 1 15 normal normal 4.057469e-04 0.9995943
3: 1 27 normal normal 7.387341e-07 0.9999993
4: 1 30 normal normal 1.959313e-04 0.9998041
5: 1 33 normal normal 1.123090e-04 0.9998877
6: 1 34 normal normal 8.422928e-04 0.9991577
response_ranger prob.suspect_ranger prob.normal_ranger
1: normal 0.306187443 0.6938126
2: normal 0.000000000 1.0000000
3: normal 0.001503821 0.9984962
4: normal 0.004442368 0.9955576
5: normal 0.004255131 0.9957449
6: normal 0.000000000 1.0000000UQ for (nested) CV - bootstrap approaches
- Simple bootstrap
- resample observations/rows of original evaluation data with replacement
- apply function (metric/statistic) to each resampled dataset
- repeat…
- Hierarchical bootstrap
- resample fold id (in this case 1:5) with replacement
- resample observations within these folds with replacement
- apply function (metric/statistic) to each resampled dataset
- repeat…
UQ for (nested) CV - hierarchical bootstrap in R
UQ for (nested) CV - bootstrap approaches
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.01122 0.02110 0.02334 0.02344 0.02568 0.03747 Min. 1st Qu. Median Mean 3rd Qu. Max.
0.008422 0.020422 0.023595 0.023605 0.026817 0.040722 UQ for (nested) CV - bootstrap approaches
- delta = AUC(ranger) - AUC(glmnet)
- quantile computation below amounts to type “percentile” in boot::boot.ci
Multiple metrics
- Usually no multiplicity correction when multiple metrics are assesed (one per performance dimension: discrimination, calibration, …)
- per performance dimension:
- define primary metric of interest
- additional secondary metrics can be investigated
Train-tune-test with single “test” model

Train-tune-test with multiple comparisons
Multiple models, multiple metrics
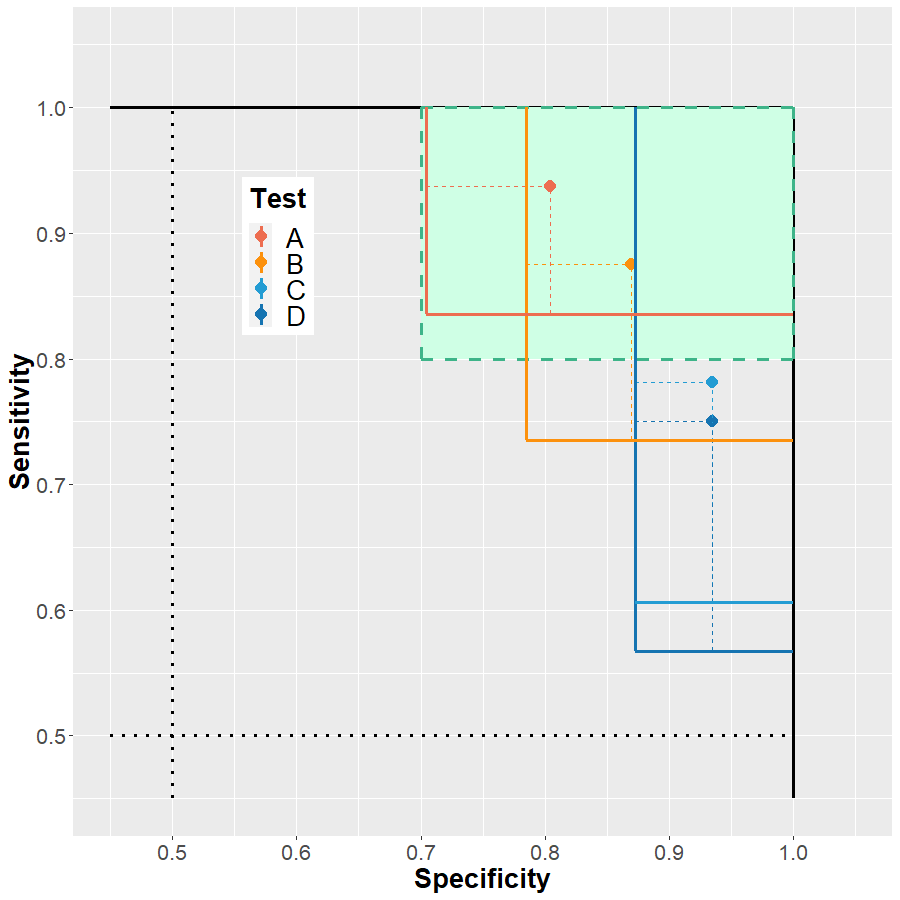
Interactive summary
Q1: What is NOT a typical goal of the statistical analysis in ML evaluation studies?
- hyperparameter optimization
- performance estimation
- hypothesis testing
- uncertainty quantification
Q1: What is NOT a typical goal of the statistical analysis in ML evaluation studies?
- hyperparameter optimization
- performance estimation
- hypothesis testing
- uncertainty quantification
Q2: Which is a valid choice of a comparator?
- another performance metric
- another prediction model
- a fixed threshold
- none
Q2: Which is a valid choice of a comparator?
- another performance metric
- another prediction model
- a fixed threshold
- none
Q3: The classical boostrap should in general NOT be used…
- for resampling / data splitting
- as part of learning algorithms
- for uncertainty quantification in simple scenarios
- for statistical anaylsis of hierarchical data
Q3: The classical boostrap should in general NOT be used…
- for resampling / data splitting
- as part of learning algorithms
- for uncertainty quantification in simple scenarios
- for statistical anaylsis of hierarchical data
Q4: Multiple models should not be assessed on the test dataset, unless…
- they are all trained by the same learning algorithm
- they have been developed by different persons/groups
- a correction for multiple comparisons is employed
- the paper will be submitted to “Nature Machine Intelligence”
Q4: Multiple models should not be assessed on the test dataset, unless…
- they are all trained by the same learning algorithm
- they have been developed by different persons/groups
- a correction for multiple comparisons is employed
- the paper will be submitted to “Nature Machine Intelligence”
Questions
Practical aspects
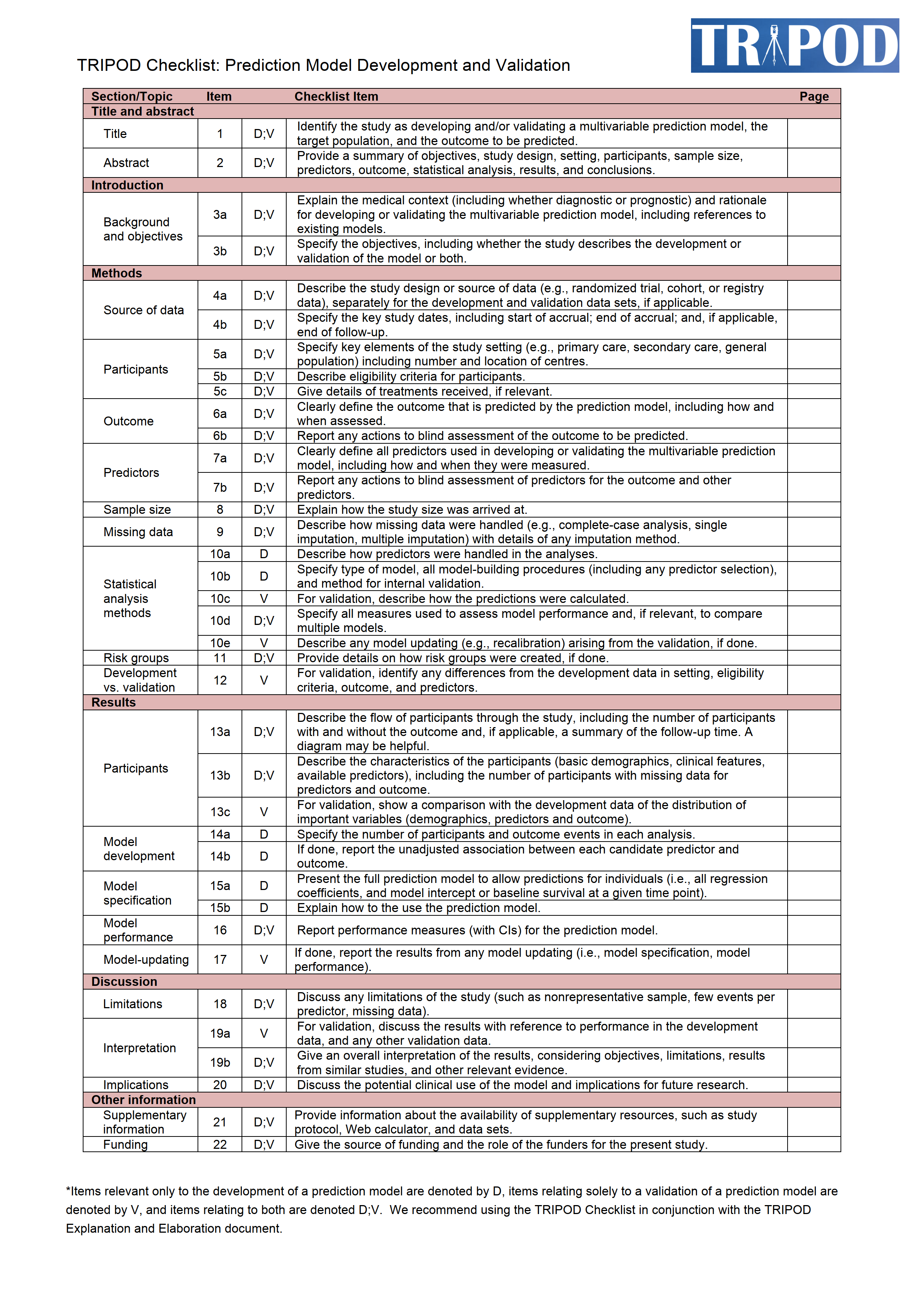
Reporting guidelines
- TRIPOD: Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement (2015)
- TRIPOD-AI (in preparation)
- REFORMS: Reporting Standards for ML-based Science (2023)
TRIPOD
Planning ahead
flowchart LR MS(Metric selection) --> SA(Statistical analysis) DS(Data splitting) --> SA SA --> R(Reporting)
- Study protocol (e.g. for prospective data aquisition)
- Statistical analysis plan
Project organization
- The “conflict of interest” of applied ML
- we want high numbers (empirical performance estimates)
- we want meaningful numbers (e.g. unbiased estimates)
- Splitting up work for development, evaluation (between persons, teams, institutions) makes things a whole lot easier
- different teams should still work together
- a good evaluation effort can support model development
Requirement analysis
- Typical requirements for ML solutions
- need for predicted probabilities
- sufficient interpretability (limited complexity)
- fast inference speed
- …
- Requirements should be assessed initially, not after performance evaluation.
- This usually requires discussion with domain experts.
Power calculation
- Power: probability to be able to demonstrate suitability of a/your (new) model/algorithm
- For evaluation on test set after training and tuning: a wide variety of methods exists (classification \(\leftrightarrow\) diagnostic accuracy studies)
- Hypothesis testing based
- Precision based, e.g. here: https://shiny.ctu.unibe.ch/app_direct/presize/
Target population/setting
- What is the target population/setting?
- Define sensible inclusion/exclusion criteria
- Helps to assemble the test set (IEC are met)
- During development, things may well be (and often are) a little “wilder”:
- Synthetic data
- Transfer learning (other population/setting)
- Data augmentation (e.g. manipulation of medical images)
- None of these is sensible in an evaluation context!
- Exception: sensitivity analyses…
Reproducibility
- Reproducibility is important but also hard (in particular in ML)
- many sources of variability
- volatile software packages
- non-deterministic learning
- Some practical considerations
- random seed(ing) (set.seed(123))
- version control (e.g. github)
- experiment handling (e.g. batchtools package)
- pipeline management (e.g. targets package)
Interactive summary
Q1: What is NOT a reporting guideline for predictive modelling studies?
- TRIPOD
- TRIPOD-AI
- REPORT-AI
- REFORMS
Q1: What is NOT a reporting guideline for predictive modelling studies?
- TRIPOD
- TRIPOD-AI
- REPORT-AI
- REFORMS
Q2: What is mostly NOT depending on the other options and should thus be decided upon initially?
- data splitting
- statistical analysis
- metric selection
- reporting
Q2: What is mostly NOT depending on the other options and should thus be decided upon initially?
- data splitting
- statistical analysis
- metric selection
- reporting
Q3: The “conflict of interest of predictive modelling” can be (partially) avoided by…
- avoiding any claims on statistical significance
- separating model development and model evaluation
- adequate & transparent study planning
- intransparent reporting
Q3: The “conflict of interest of predictive modelling” can be (partially) avoided by…
- avoiding any claims on statistical significance
- separating model development and model evaluation
- adequate & transparent study planning
- intransparent reporting
Q4: Power analyses…
- can help to avoid conducting unreasonably large studies
- are independent of the chosen evaluation metric
- are only useful if hypothesis testing is planned for evaluation
- should be reported according to the TRIPOD statement
Q4: Power analyses…
- can help to avoid conducting unreasonably large studies
- are independent of the chosen evaluation metric
- are only useful if hypothesis testing is planned for evaluation
- should be reported according to the TRIPOD statement
Q5: For prospective studies, additional care is required…
- to avoid leakage
- to avoid statistically significant results
- to end up with an adequate test set
- to define the performance metric(s)
Q5: For prospective studies, additional care is required…
- to avoid leakage
- to avoid statistically significant results
- to end up with an adequate test set
- to define the performance metric(s)
Questions
Wrap-up
Learnings
- Several pitfalls are very prevalent in applied ML.
- Overfitting and selection-induced bias cause overoptimistic evaluation results.
- Performance metric(s) should be chosen deliberately.
- Multiple dimensions can/should to be considered: discrimination, calibration, fairness, …
- Calibration is usually harder to achieve than discrimination.
- Data splitting is mandatory to avoid overoptimism.
- Large(r) samples: train-tune-test \(\rightarrow\) simple(r).
- Fewe(r) samples: (nested) CV (+test) \(\rightarrow\) (more) complex.
- Statistical analysis depends on metric choice and data splitting approach.
- Bootstrap can deal with any metric and requires minimal assumptions.
- Plan ahead (requirement analysis) and involve important stakeholders.
- Evaluation should guide/support development.
Contact
-
- Feedback
- Further questions
- Collaboration requests
- @MaxWestphal89
- Get/stay in touch
- https://github.com/maxwestphal/evaluation_in_supervised_ml_cen_2023/issues
- Issues with slides/code
Have 10 minutes to spare?
- In a current research project, we develop an interactive expert system for methodological guidance in applied ML (beyond model evaluation).
- We have setup a short survey to assess user needs and requirements.
- Thank you for your time!
https://websites.fraunhofer.de/mlguide-user-survey/index.php/578345/newtest/Y?G00Q23=CEN23
Thank you for your participation! Have a great conference!
Max Westphal & Rieke Alpers - Model and Algorithm Evaluation in Supervised Machine Learning - Short Course at CEN 2023 Conference - 2023-09-03