Code
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionCode
library(ggplot2)library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(ggplot2)knitr::opts_chunk$set(cache=FALSE)owntheme <- theme(legend.position = "bottom",
title=element_text(face="bold", size=24),
axis.title = element_text(face="bold", size=20),
legend.text = element_text(face="plain", size=20),
legend.title = element_blank(),
axis.text = element_text(size=16)) ## set input dir, needs to be equal to out.dir from
## cases_simstudy_lfc.R and cases_simstudy_roc.R:
in.dir <- file.path("E:/cases_simstudy/cases_sim_results")For local reproduction, the input directory potentially needs to be adapted!
data_lfc <- readr::read_csv2(file.path(in.dir, "cases_SIM_LFC.csv"))ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.Rows: 5600000 Columns: 61
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (10): problem, algorithm, cortype, contrast, alternative, adjustment, tr...
dbl (50): job.id, nrep, n, prev, m, se, sp, L, rhose, rhosp, benchmark, alph...
lgl (1): random
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.data_lfc <- data_lfc %>%
mutate(Adjustment = recode_factor(
interaction(adjustment, pars),
"none.list()" = "none",
"bonferroni.list()"="Bonferroni",
"maxt.list()" = "maxT",
"mbeta.list()" = "mBeta",
"bootstrap.list(type='wild')" = "Bootstrap (wild)",
"bootstrap.list()" = "Bootstrap (pairs)")
)
dim(data_lfc)[1] 5600000 62data_lfc <- data_lfc %>%
filter(cortype == "equi") %>%
filter(rhose == 0.5) %>%
filter(transformation == "none")
dim(data_lfc)[1] 2400000 62data_lfc %>%
filter(n<1000, prev==0.25, se==0.8, m==10) %>%
group_by(Adjustment, n, prev, se, m) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'se'. You can
override using the `.groups` argument.Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.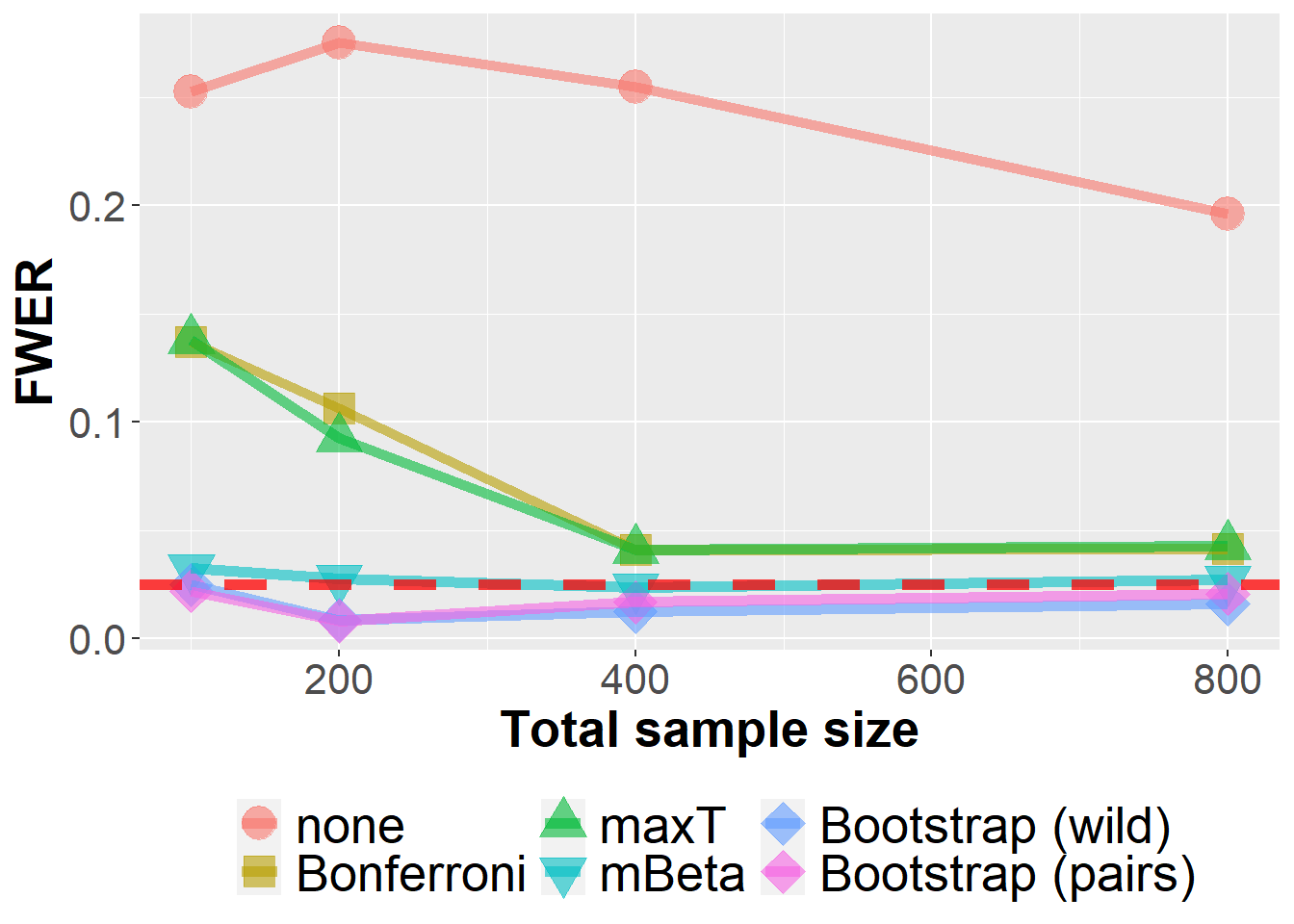
data_lfc %>%
filter(n<1000, prev==0.25, se==0.8, m==10) %>%
group_by(Adjustment, n, prev, se, m) %>%
summarize(nsim=n(), rr = mean(`0.05` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'se'. You can
override using the `.groups` argument.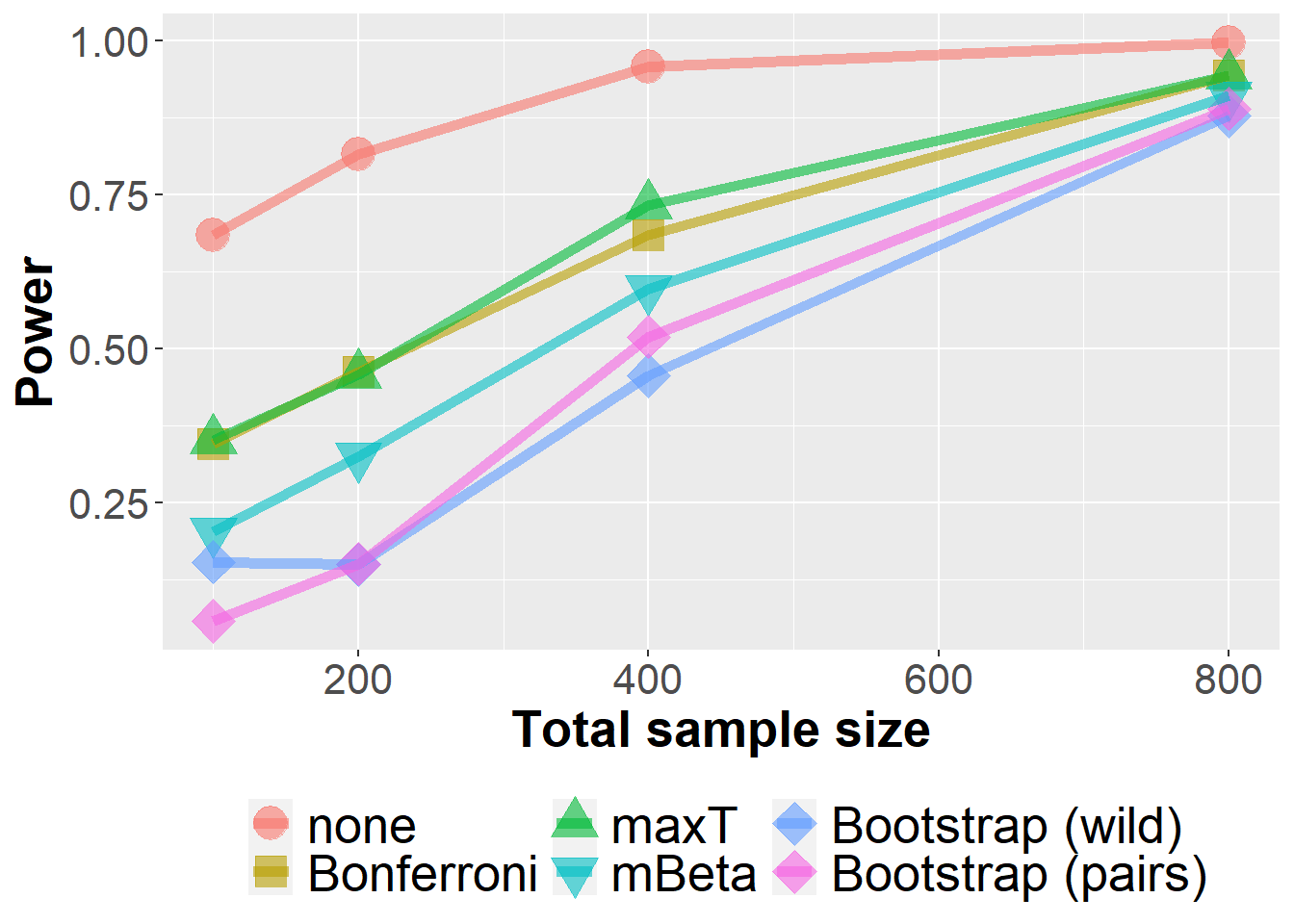
data_lfc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, se, m) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
facet_wrap(prev+se ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "se = " ~ .(se) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'se'. You can
override using the `.groups` argument.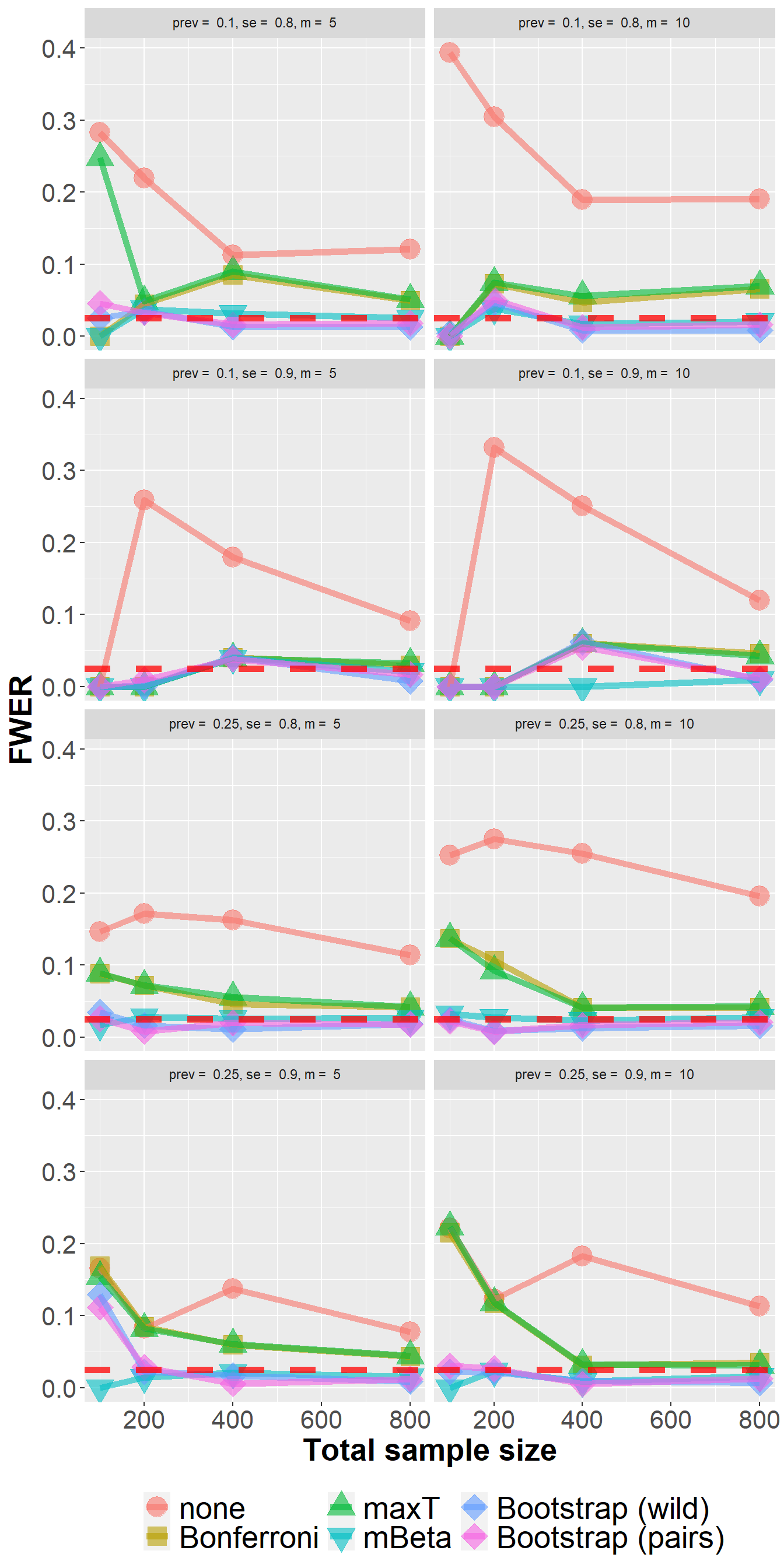
data_lfc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, se, m) %>%
summarize(nsim=n(), rr = mean(`0.05` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme +
facet_wrap(prev+se ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "se = " ~ .(se) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'se'. You can
override using the `.groups` argument.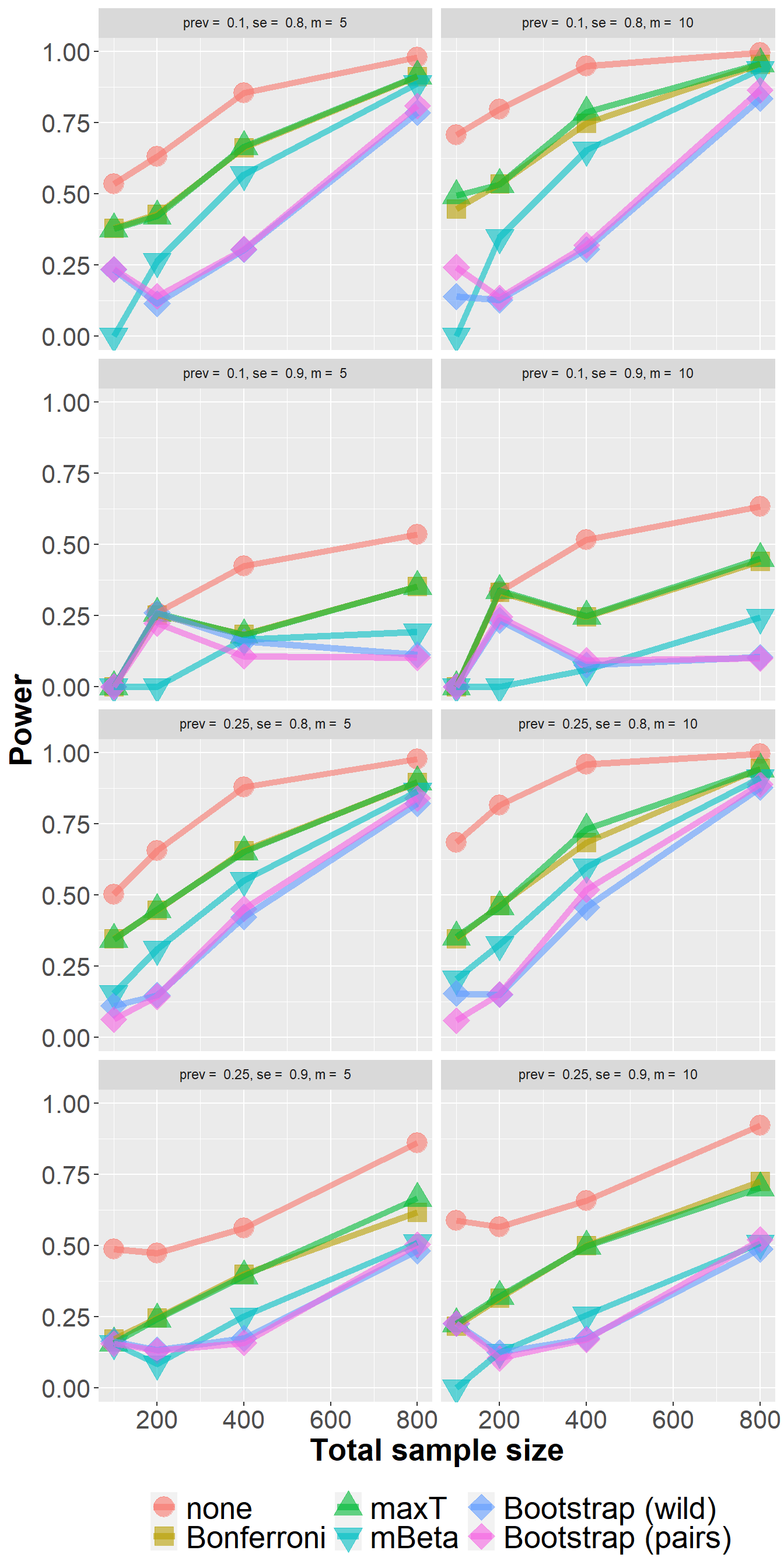
data_lfc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, se, m) %>%
summarize(nsim=n(), rr = mean(`0.1` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme +
facet_wrap(prev+se ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "se = " ~ .(se) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'se'. You can
override using the `.groups` argument.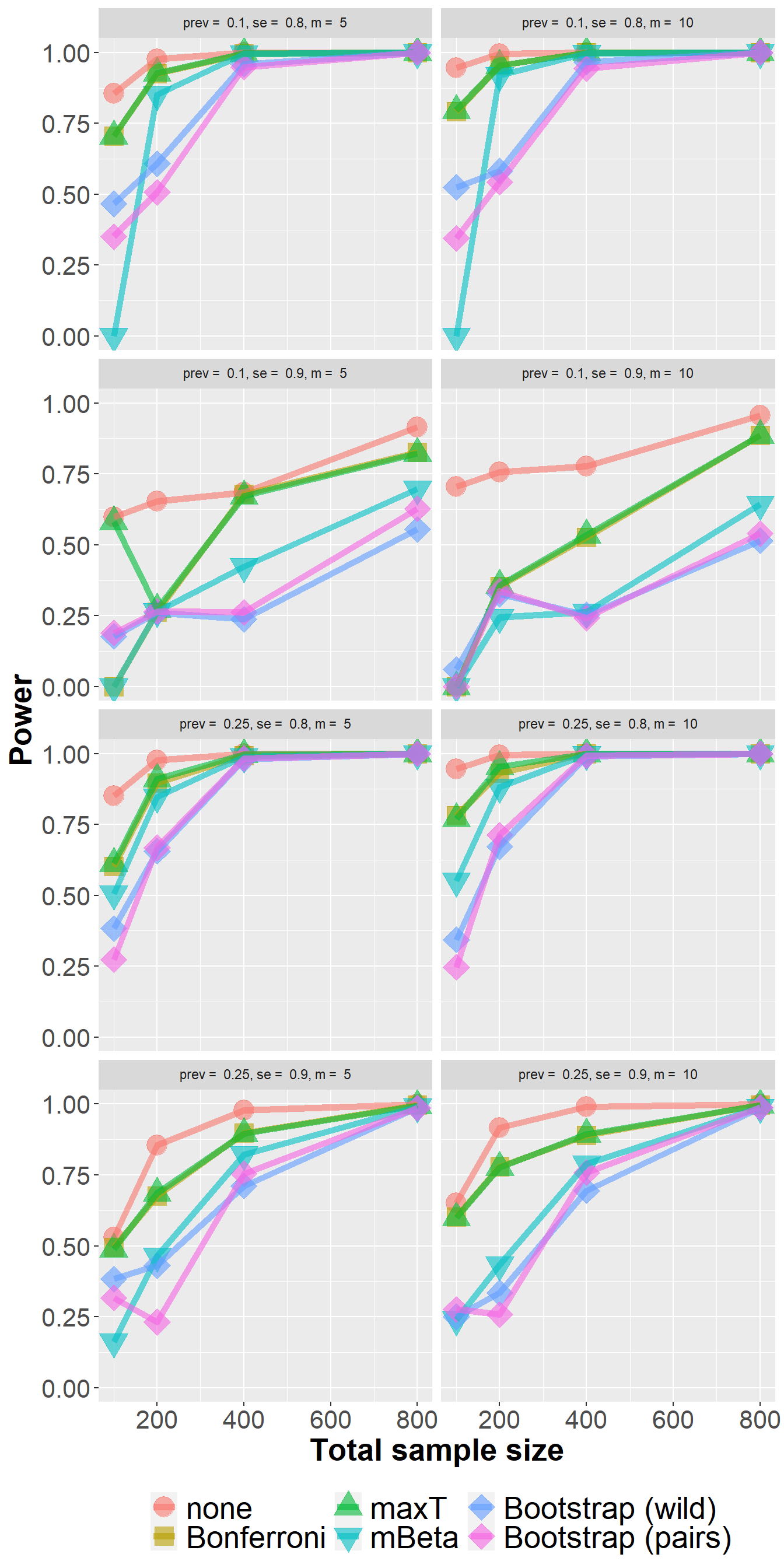
data_roc <- readr::read_csv2(file.path(in.dir, "cases_SIM_ROC.csv"))ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.Rows: 8400000 Columns: 61
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (10): problem, algorithm, auc, contrast, alternative, adjustment, transf...
dbl (50): job.id, nrep, n, prev, m, rhose, e, k, delta, rhosp, benchmark, al...
lgl (1): random
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.data_roc <- data_roc %>%
mutate(Adjustment = recode_factor(
interaction(adjustment, pars),
"none.list()" = "none",
"bonferroni.list()"="Bonferroni",
"maxt.list()" = "maxT",
"mbeta.list()" = "mBeta",
"bootstrap.list(type='wild')" = "Bootstrap (wild)",
"bootstrap.list()" = "Bootstrap (pairs)")
)
dim(data_roc)[1] 8400000 62data_roc <- data_roc %>%
filter(rhose == 0.5) %>%
filter(auc == "seq(0.85, 0.90, length.out = 5)") %>%
filter(transformation == "none")
dim(data_roc)[1] 1200000 62data_roc %>%
filter(n<1000, prev==0.25, m==10) %>%
group_by(Adjustment, n, prev, m) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme `summarise()` has grouped output by 'Adjustment', 'n', 'prev'. You can override
using the `.groups` argument.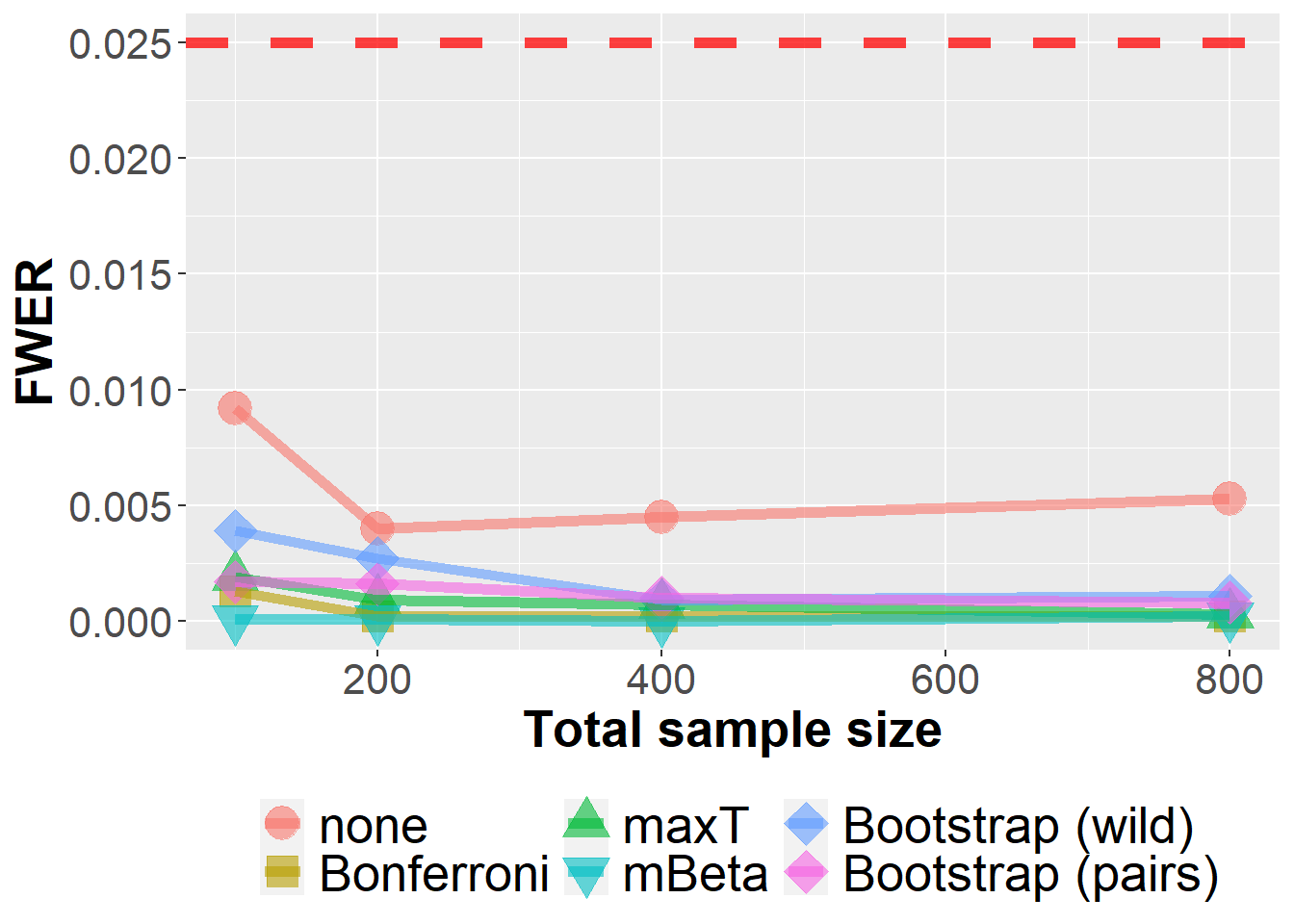
data_roc %>%
filter(n<1000, prev==0.25, m==10) %>%
group_by(Adjustment, n, prev, m) %>%
summarize(nsim=n(), rr = mean(`0.1` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme `summarise()` has grouped output by 'Adjustment', 'n', 'prev'. You can override
using the `.groups` argument.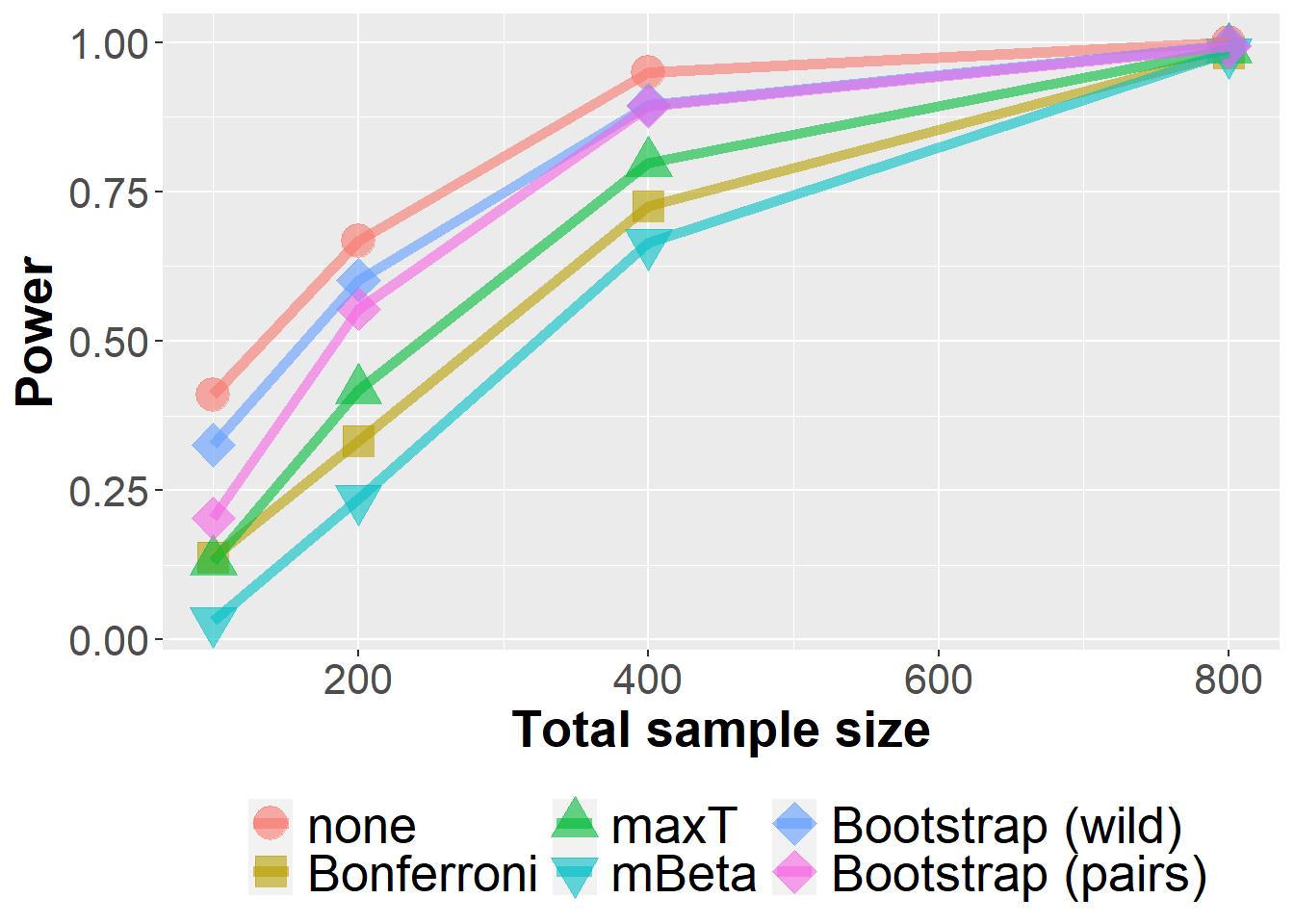
data_roc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, m) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
facet_wrap(prev ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev'. You can override
using the `.groups` argument.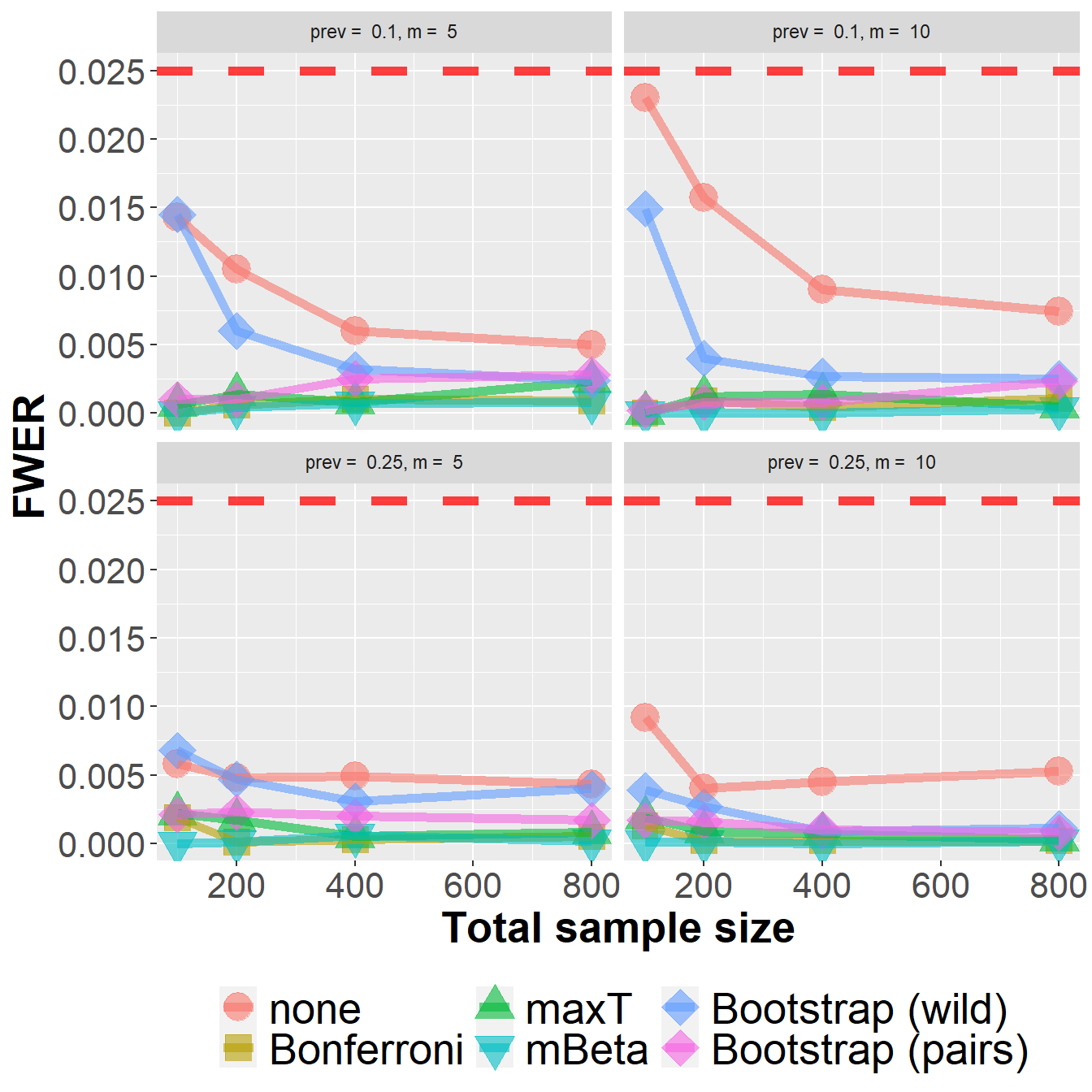
data_roc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, m) %>%
summarize(nsim=n(), rr = mean(`0.1` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme +
facet_wrap(prev ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev'. You can override
using the `.groups` argument.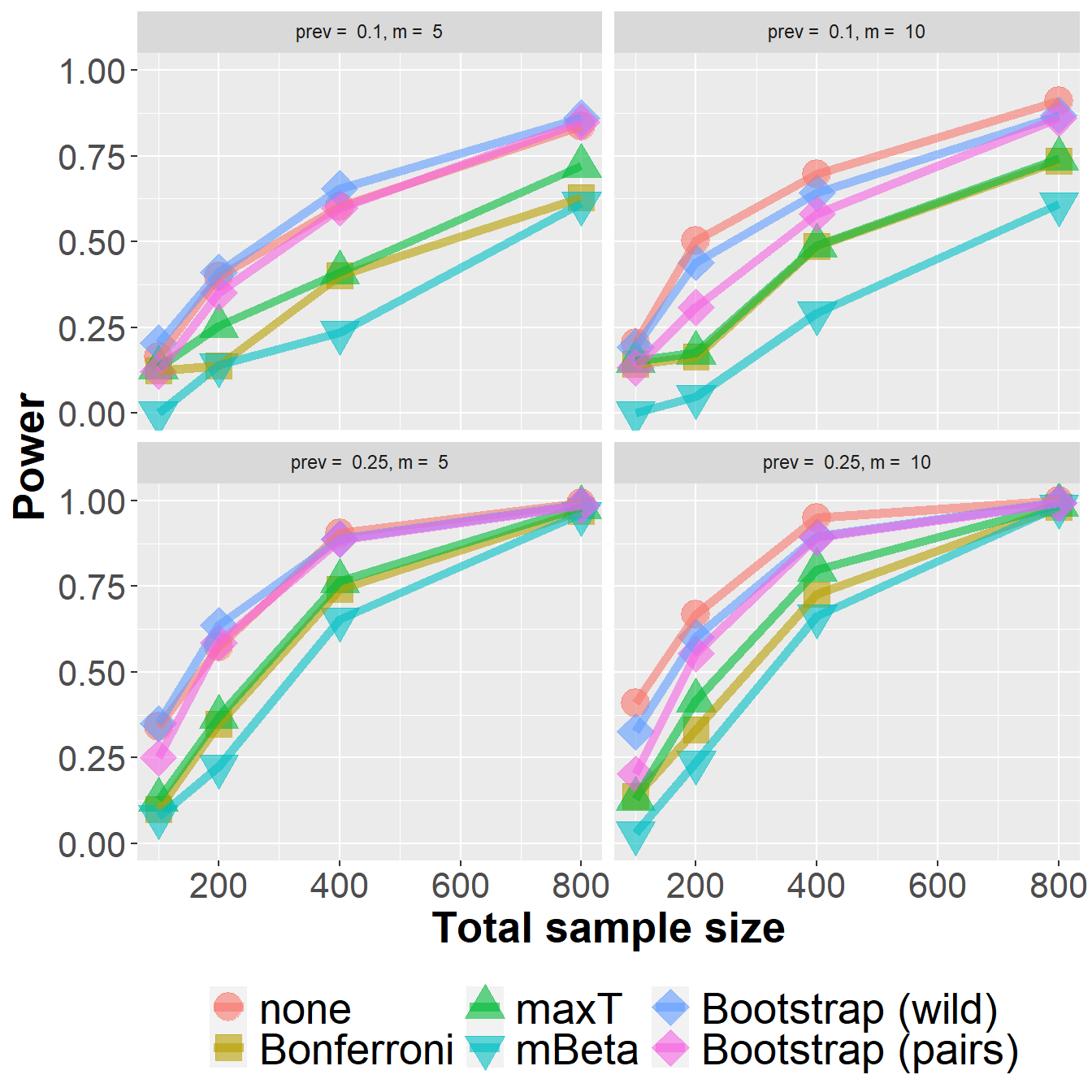
data_roc %>%
filter(n<1000) %>%
group_by(Adjustment, n, prev, m) %>%
summarize(nsim=n(), rr = mean(`0.05` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment)) +
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("Power") +
scale_shape_manual(values = c(21, 22, 24, 25, 23, 23)) +
owntheme +
facet_wrap(prev ~ m, ncol=2,
labeller = label_bquote("prev = " ~ .(prev) * ", " * "m = " ~ .(m)))`summarise()` has grouped output by 'Adjustment', 'n', 'prev'. You can override
using the `.groups` argument.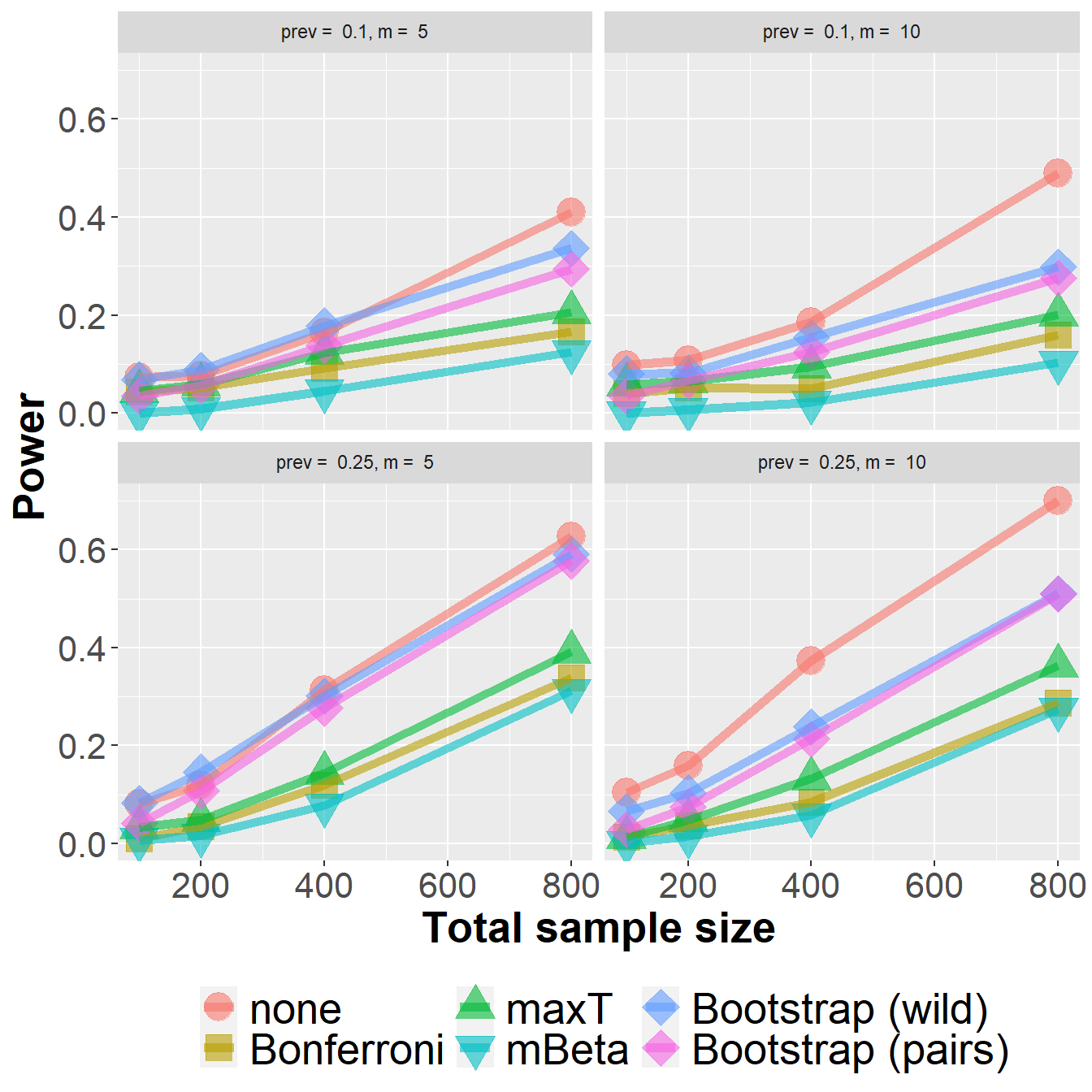
in.dir <- file.path("E:/cases_SIM/cases_SIM_results") # TODO: remove
data_sens_lfc <- readr::read_csv2(file.path(in.dir, "cases_SIM_SENS_lfc.csv"))ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.Rows: 400000 Columns: 61
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (10): problem, algorithm, cortype, contrast, alternative, adjustment, tr...
dbl (50): job.id, nrep, n, prev, m, se, sp, L, rhose, rhosp, benchmark, alph...
lgl (1): random
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.data_sens_lfc <- data_sens_lfc %>%
mutate(Adjustment = recode_factor(
interaction(adjustment, pars, drop = TRUE),
"none.list()" = "none",
"bonferroni.list()"="Bonferroni",
"maxt.list()" = "maxT",
"mbeta.list()" = "mBeta (lfc_pr=1)",
"mbeta.list(lfc_pr=0.5)" = "mBeta (lfc_pr=0.5)",
"mbeta.list(lfc_pr=0)" = "mBeta (lfc_pr=0)",
"bootstrap.list(type='wild')" = "Bootstrap (wild)",
"bootstrap.list()" = "Bootstrap (pairs)")
)
dim(data_sens_lfc)[1] 400000 62data_sens_roc <- readr::read_csv2(file.path(in.dir, "cases_SIM_SENS_roc.csv"))ℹ Using "','" as decimal and "'.'" as grouping mark. Use `read_delim()` for more control.Rows: 800000 Columns: 62
── Column specification ────────────────────────────────────────────────────────
Delimiter: ";"
chr (11): problem, algorithm, auc, dist, contrast, alternative, adjustment, ...
dbl (50): job.id, nrep, n, prev, m, rhose, e, k, delta, rhosp, benchmark, al...
lgl (1): random
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.data_sens_roc <- data_sens_roc %>%
mutate(Adjustment = recode_factor(
interaction(adjustment, pars, drop = TRUE),
"none.list()" = "none",
"bonferroni.list()"="Bonferroni",
"maxt.list()" = "maxT",
"mbeta.list()" = "mBeta (lfc_pr=1)",
"mbeta.list(lfc_pr=0.5)" = "mBeta (lfc_pr=0.5)",
"mbeta.list(lfc_pr=0)" = "mBeta (lfc_pr=0)",
"bootstrap.list(type='wild')" = "Bootstrap (wild)",
"bootstrap.list()" = "Bootstrap (pairs)")
)
dim(data_sens_roc)[1] 800000 63In this sensitvity analysis, the data generating ROC model was altered to the a bi-exponential model instead of the default bi-normal model.
data_sens_roc %>%
filter(n<1000) %>%
filter(!(Adjustment %in% c("mBeta (lfc_pr=0.5)", "mBeta (lfc_pr=0)"))) %>%
group_by(Adjustment, n, prev, m, dist) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=dist, fill=dist, pch=dist))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
facet_wrap(~ Adjustment, ncol=2)`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'm'. You can
override using the `.groups` argument.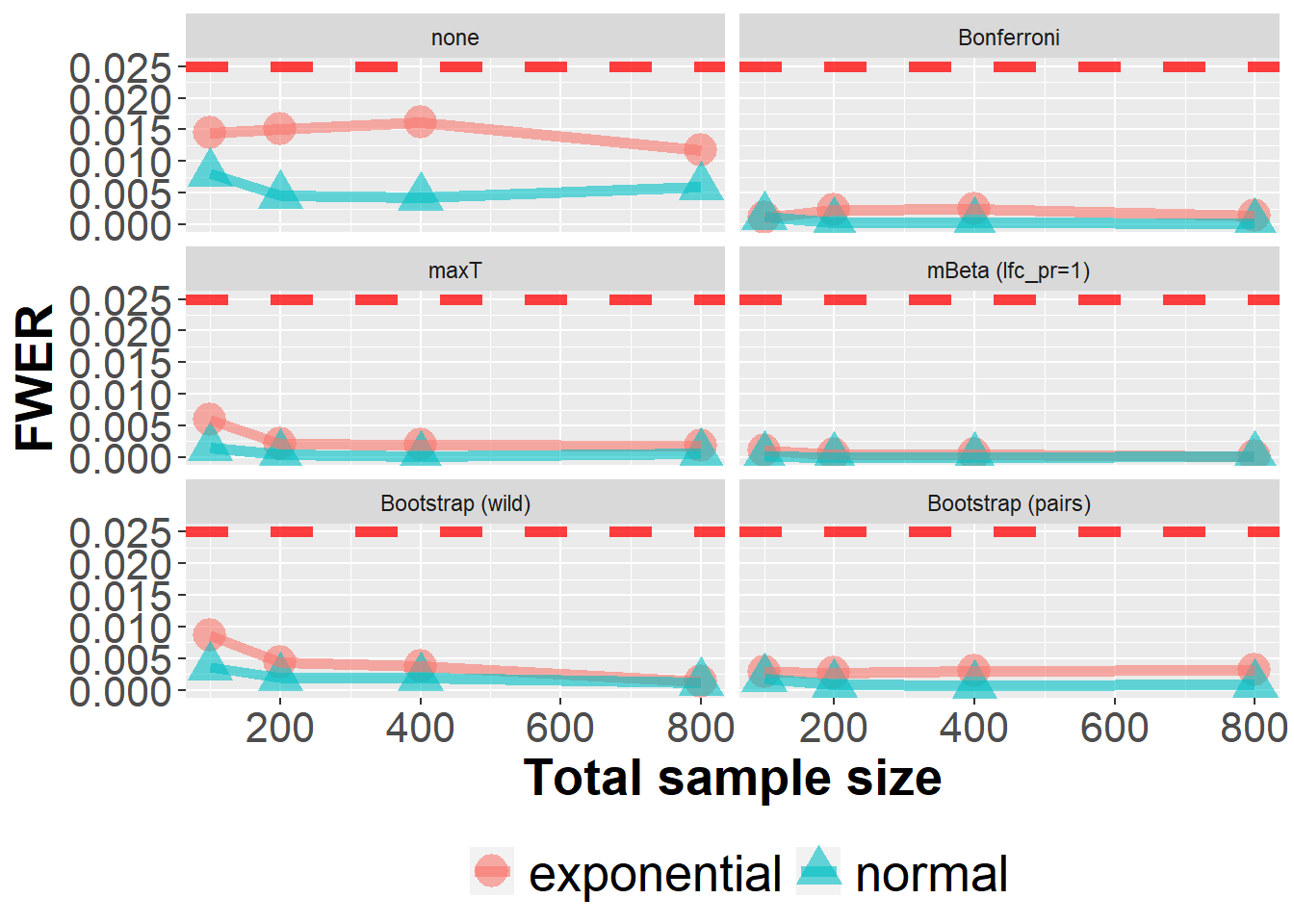
The next plot presents the same data slighty differently.
data_sens_roc %>%
filter(n<1000) %>%
filter(!(Adjustment %in% c("mBeta (lfc_pr=0.5)", "mBeta (lfc_pr=0)"))) %>%
group_by(Adjustment, n, prev, m, dist) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
facet_wrap(~ dist, ncol=1)`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'm'. You can
override using the `.groups` argument.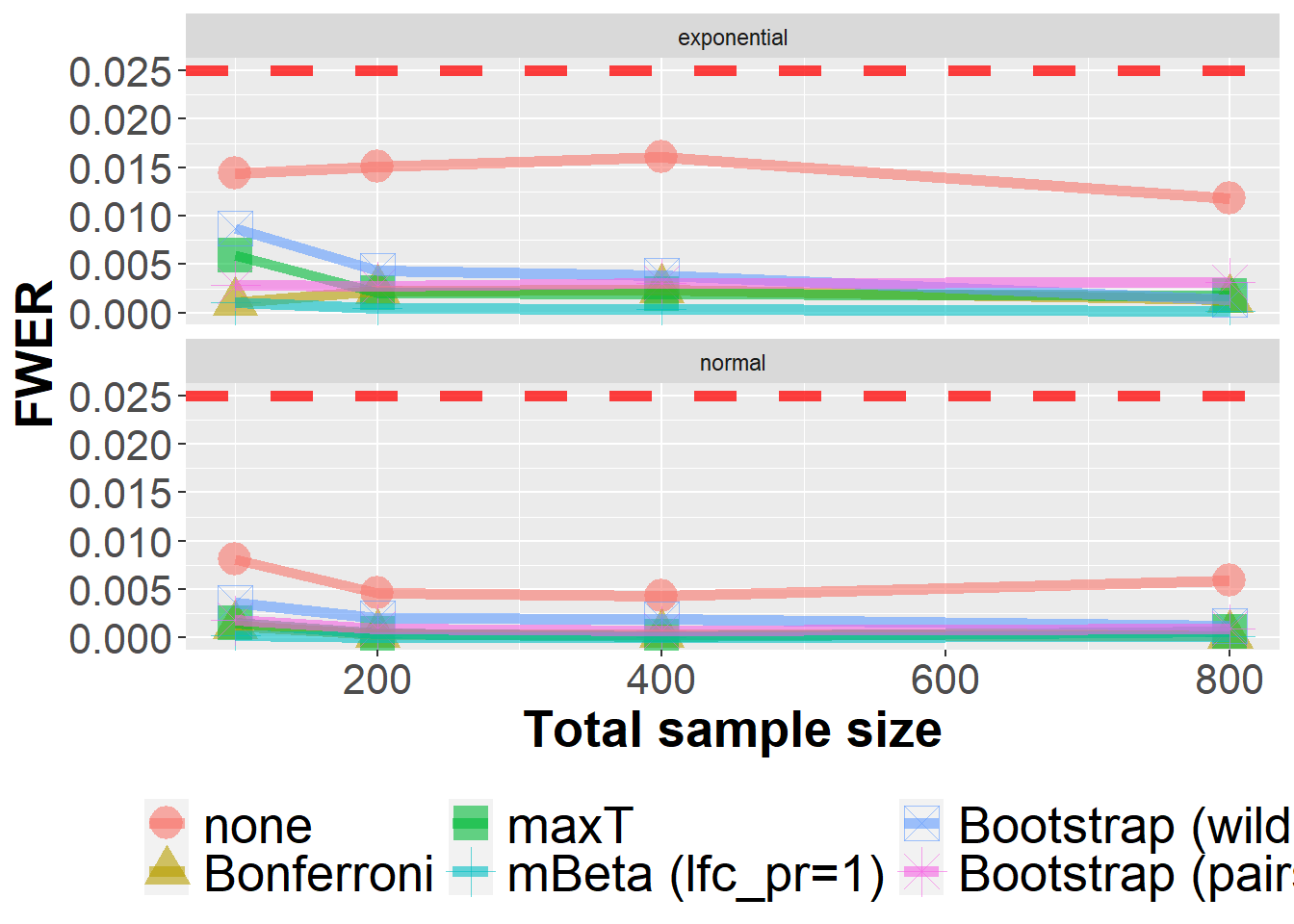
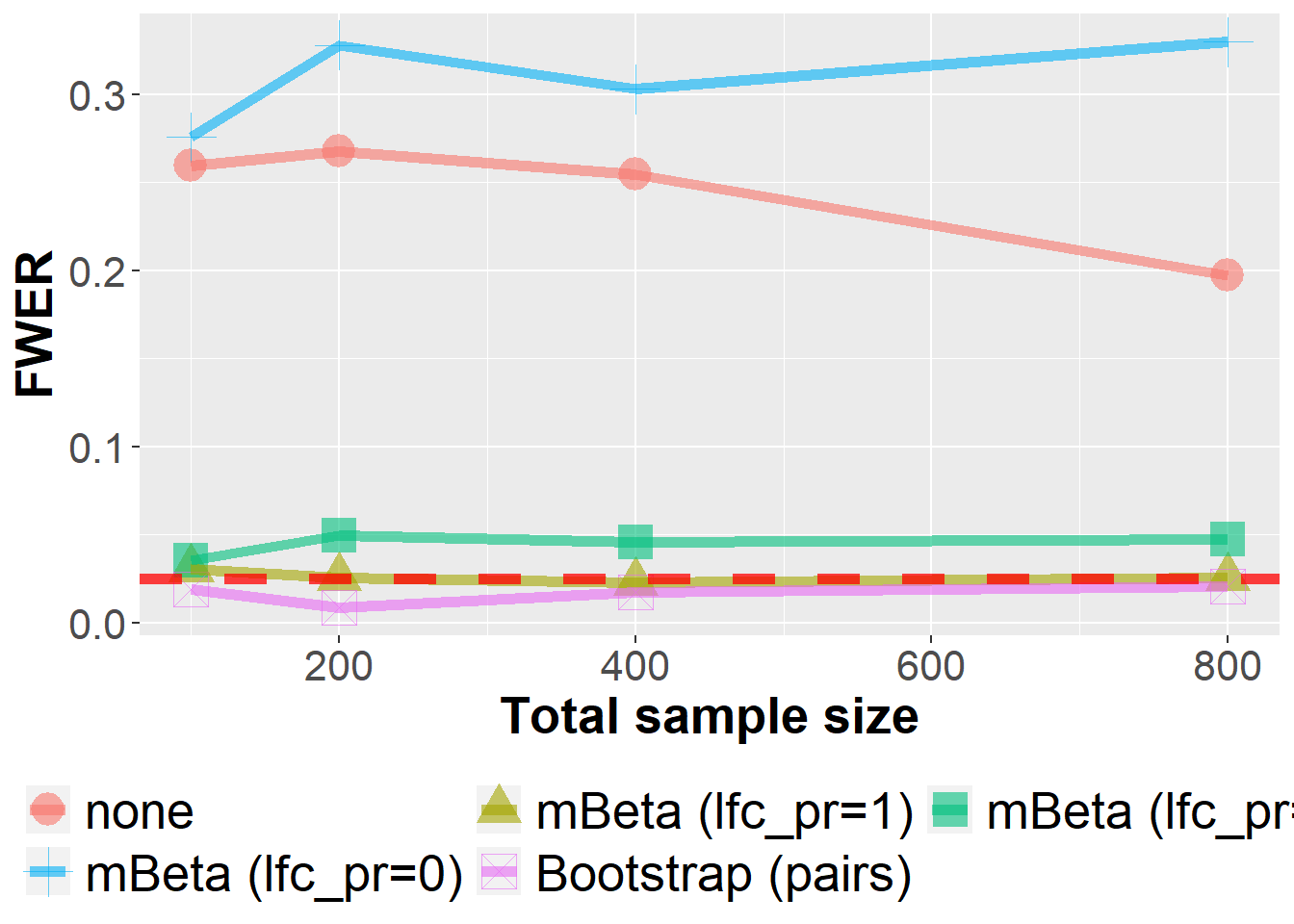
The mBeta adjustment is a heuristic approach which builds on a Bayesian multivariate Beta-Binomial model. In this sensitivity analysis, the influence of the “lfc_pr’ hyperparameter is investigated. It lies in the unit interval and controls if the prior distribution is solely a least-favorable parameter configuration (lfc_pr=1, default), a mixture distribution (lfc_pr=0.5) or solely a uniform distribution (lfc_pr=0). As we can see below, this parameter has a noticeable influence on operating characteristics as the FWER.
data_sens_roc %>%
filter(n<1000, dist=="normal") %>%
filter(Adjustment %in% c("none", "mBeta (lfc_pr=1)", "mBeta (lfc_pr=0.5)", "mBeta (lfc_pr=0)", "Bootstrap (pairs)")) %>%
group_by(Adjustment, n, prev, m, dist) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
guides(fill=guide_legend(nrow=2, byrow=TRUE),
col=guide_legend(nrow=2, byrow=TRUE),
pch=guide_legend(nrow=2, byrow=TRUE))`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'm'. You can
override using the `.groups` argument.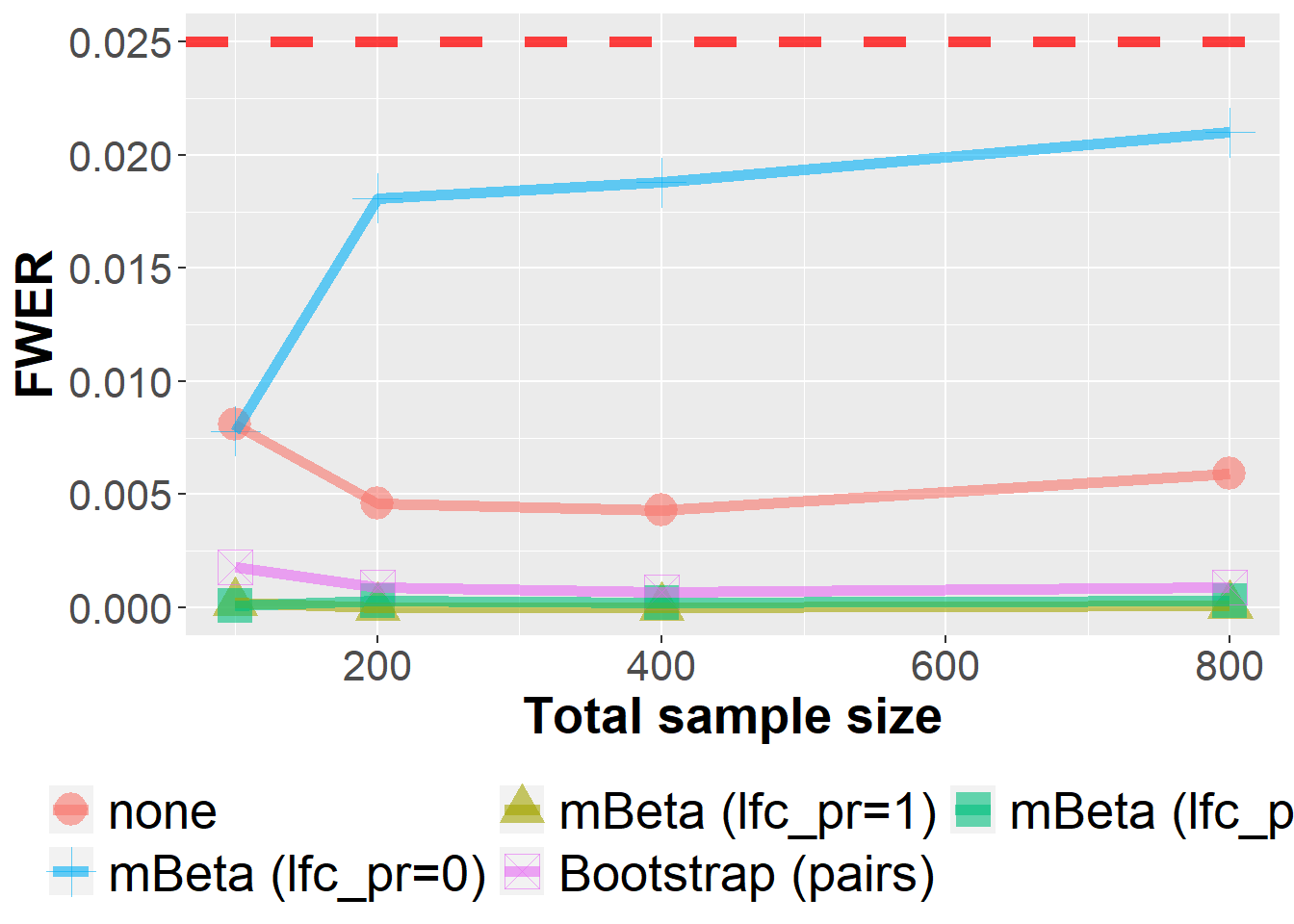
data_sens_lfc %>%
filter(n<1000, se==0.8) %>%
filter(Adjustment %in% c("none", "mBeta (lfc_pr=1)", "mBeta (lfc_pr=0.5)", "mBeta (lfc_pr=0)", "Bootstrap (pairs)")) %>%
group_by(Adjustment, n, prev, m, se) %>%
summarize(nsim=n(), rr = mean(`0` > 0)) %>%
ggplot(aes(n, rr, col=Adjustment, fill=Adjustment, pch=Adjustment))+
geom_point(size=6, alpha=0.6) +
geom_line(size=2, alpha=0.6) +
xlab("Total sample size") +
ylab("FWER") +
geom_hline(yintercept=0.025, col="red", lwd=2, lty=2, alpha=0.75) +
owntheme +
guides(fill=guide_legend(nrow=2, byrow=TRUE),
col=guide_legend(nrow=2, byrow=TRUE),
pch=guide_legend(nrow=2, byrow=TRUE))`summarise()` has grouped output by 'Adjustment', 'n', 'prev', 'm'. You can
override using the `.groups` argument.